Chromosome-level genome assembly of largemouth bass (Micropterus salmoides) using PacBio and Hi-C technologies
- PMID: 35933561
- PMCID: PMC9357066
- DOI: 10.1038/s41597-022-01601-1
Chromosome-level genome assembly of largemouth bass (Micropterus salmoides) using PacBio and Hi-C technologies
Abstract
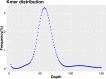
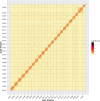
The largemouth bass (Micropterus salmoides) has become a cosmopolitan species due to its widespread introduction as game or domesticated fish. Here a high-quality chromosome-level reference genome of M. salmoides was produced by combining Illumina paired-end sequencing, PacBio single molecule sequencing technique (SMRT) and High-through chromosome conformation capture (Hi-C) technologies. Ultimately, the genome was assembled into 844.88 Mb with a contig N50 of 15.68 Mb and scaffold N50 length of 35.77 Mb. About 99.9% assembly genome sequences (844.00 Mb) could be anchored to 23 chromosomes, and 98.03% assembly genome sequences could be ordered and directed. The genome contained 38.19% repeat sequences and 2693 noncoding RNAs. A total of 26,370 protein-coding genes from 3415 gene families were predicted, of which 97.69% were functionally annotated. The high-quality genome assembly will be a fundamental resource to study and understand how M. salmoides adapt to novel and changing environments around the world, and also be expected to contribute to the genetic breeding and other research.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Bai J, Dijar L-C, Quan Y, Liang S. Taxonomic status and genetic diversity of cultured largemouth bass Micropterus salmoides in China. Aquaculture. 2008;278:27–30. doi: 10.1016/j.aquaculture.2008.03.016. - DOI
-
- García-Berthou E, et al. Introduction pathways and establishment rates of invasive aquatic species in Europe. Can. J. Fish. Aquat. Sci. 2005;62:453–463. doi: 10.1139/f05-017. - DOI
-
- García-Berthou, E. Ontogenetic diet shifts and interrupted piscivoryin introduced largemouth bass (Micropterus salmoides). Int. Rev. Hydrobiol. 87, 353–363, 10.1002/1522-2632(200207)87:4%3C353::AID-IROH353%3E3.0.CO;2-N (2002).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

