Living Biomaterials to Engineer Hematopoietic Stem Cell Niches
- PMID: 35933595
- PMCID: PMC11469072
- DOI: 10.1002/adhm.202200964
Living Biomaterials to Engineer Hematopoietic Stem Cell Niches
Abstract
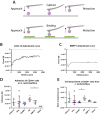
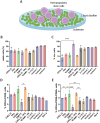
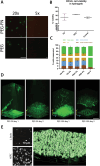
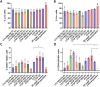
Living biointerfaces are a new class of biomaterials combining living cells and polymeric matrices that can act as biologically active and instructive materials that host and provide signals to surrounding cells. Here, living biomaterials based on Lactococcus lactis to control hematopoietic stem cells in 2D surfaces and 3D hydrogels are introduced. L. lactis is modified to express C-X-C motif chemokine ligand 12 (CXCL12), thrombopoietin (TPO), vascular cell adhesion protein 1 (VCAM1), and the 7th-10th type III domains of human plasma fibronectin (FN III7-10 ), in an attempt to mimic ex vivo the conditions of the human bone marrow. These results suggest that living biomaterials that incorporate bacteria expressing recombinant CXCL12, TPO, VCAM1, and FN in both 2D systems direct hematopoietic stem and progenitor cells (HSPCs)-bacteria interaction, and in 3D using hydrogels functionalized with full-length human plasma fibronectin allow for a notable expansion of the CD34+ /CD38- /CD90+ HSPC population compared to the initial population. These results provide a strong evidence based on data that suggest the possibility of using living materials based on genetically engineered bacteria for the ex-vivo expansion of HSPC with eventual practical clinical applications in HSPCs transplantation for hematological disorders.
Keywords: cell engineering; genetic engineering; hematopoiesis; living materials; microenvironment engineering; stem cells; synthetic biology.
© 2022 The Authors. Advanced Healthcare Materials published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Thrombopoietin enhances hematopoietic stem and progenitor cell homing by impeding matrix metalloproteinase 9 expression.Stem Cells Transl Med. 2020 Jun;9(6):661-673. doi: 10.1002/sctm.19-0220. Epub 2020 Mar 3. Stem Cells Transl Med. 2020. PMID: 32125099 Free PMC article.
-
Regulation of hematopoietic stem cell behavior by the nanostructured presentation of extracellular matrix components.PLoS One. 2013;8(2):e54778. doi: 10.1371/journal.pone.0054778. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405094 Free PMC article.
-
Thrombopoietin promotes adhesion of primitive human hemopoietic cells to fibronectin and vascular cell adhesion molecule-1: role of activation of very late antigen (VLA)-4 and VLA-5.J Immunol. 1997 Aug 15;159(4):1961-9. J Immunol. 1997. PMID: 9257862
-
Rebuilding the hematopoietic stem cell niche: Recent developments and future prospects.Acta Biomater. 2021 Sep 15;132:129-148. doi: 10.1016/j.actbio.2021.03.061. Epub 2021 Apr 1. Acta Biomater. 2021. PMID: 33813090 Review.
-
Current Developments in Mobilization of Hematopoietic Stem and Progenitor Cells and Their Interaction with Niches in Bone Marrow.Transfus Med Hemother. 2017 Jun;44(3):151-164. doi: 10.1159/000477262. Epub 2017 May 29. Transfus Med Hemother. 2017. PMID: 28626366 Free PMC article. Review.
Cited by
-
Intervertebral disc degeneration-Current therapeutic options and challenges.Front Public Health. 2023 Jul 6;11:1156749. doi: 10.3389/fpubh.2023.1156749. eCollection 2023. Front Public Health. 2023. PMID: 37483952 Free PMC article. Review.
-
Cell-microsphere based living microhybrids for osteogenesis regulating to boosting biomineralization.Regen Biomater. 2024 Oct 29;11:rbae125. doi: 10.1093/rb/rbae125. eCollection 2024. Regen Biomater. 2024. PMID: 39569077 Free PMC article.
-
Transcriptional regulation of living materials via extracellular electron transfer.Nat Chem Biol. 2024 Oct;20(10):1329-1340. doi: 10.1038/s41589-024-01628-y. Epub 2024 May 23. Nat Chem Biol. 2024. PMID: 38783133
-
Thymol as a Component of Chitosan Systems-Several New Applications in Medicine: A Comprehensive Review.Plants (Basel). 2024 Jan 25;13(3):362. doi: 10.3390/plants13030362. Plants (Basel). 2024. PMID: 38337895 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

