Human coronaviruses: The emergence of SARS-CoV-2 and management of COVID-19
- PMID: 35934258
- PMCID: PMC9351210
- DOI: 10.1016/j.virusres.2022.198882
Human coronaviruses: The emergence of SARS-CoV-2 and management of COVID-19
Abstract
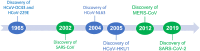
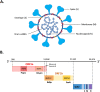
To date, a total of seven human coronaviruses (HCoVs) have been identified, all of which are important respiratory pathogens. Recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), has led to a global pandemic causing millions of infections and deaths. Here, we summarize the discovery and fundamental virology of HCoVs, discuss their zoonotic transmission and highlight the weak species barrier of SARS-CoV-2. We also discuss the possible origins of SARS-CoV-2 variants of concern identified to date and discuss the experimental challenges in characterizing mutations of interest and propose methods to circumvent them. As the COVID-19 treatment and prevention landscape rapidly evolves, we summarize current therapeutics and vaccines, and their implications on SARS-CoV-2 variants. Finally, we explore how interspecies transmission of SARS-CoV-2 may drive the emergence of novel strains, how disease severity may evolve and how COVID-19 will likely continue to burden healthcare systems globally.
Keywords: COVID-19; Cross-species transmission; Human coronaviruses; Long COVID; SARS-CoV-2; Vaccines; Variants of concern.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): A review of the current global status.J Infect Public Health. 2020 Nov;13(11):1601-1610. doi: 10.1016/j.jiph.2020.07.011. Epub 2020 Aug 4. J Infect Public Health. 2020. PMID: 32778421 Free PMC article.
-
The novel zoonotic COVID-19 pandemic: An expected global health concern.J Infect Dev Ctries. 2020 Mar 31;14(3):254-264. doi: 10.3855/jidc.12671. J Infect Dev Ctries. 2020. PMID: 32235085
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
Zoonotic origins of human coronaviruses.Int J Biol Sci. 2020 Mar 15;16(10):1686-1697. doi: 10.7150/ijbs.45472. eCollection 2020. Int J Biol Sci. 2020. PMID: 32226286 Free PMC article. Review.
-
Emergence, Transmission, and Potential Therapeutic Targets for the COVID-19 Pandemic Associated with the SARS-CoV-2.Cell Physiol Biochem. 2020 Aug 25;54(4):767-790. doi: 10.33594/000000254. Cell Physiol Biochem. 2020. PMID: 32830930
Cited by
-
COVID-19 - multisystem disease.Rom J Morphol Embryol. 2025 Jan-Mar;66(1):61-67. doi: 10.47162/RJME.66.1.05. Rom J Morphol Embryol. 2025. PMID: 40384192 Free PMC article. Review.
-
Hotspot residues and resistance mutations in the nirmatrelvir-binding site of SARS-CoV-2 main protease: Design, identification, and correlation with globally circulating viral genomes.Biochem Biophys Res Commun. 2022 Nov 12;629:54-60. doi: 10.1016/j.bbrc.2022.09.010. Epub 2022 Sep 7. Biochem Biophys Res Commun. 2022. PMID: 36113178 Free PMC article.
-
Discovery of a nasal spray steroid, tixocortol, as an inhibitor of SARS-CoV-2 main protease and viral replication.RSC Med Chem. 2024 Sep 27;15(12):4193-205. doi: 10.1039/d4md00454j. Online ahead of print. RSC Med Chem. 2024. PMID: 39371432 Free PMC article.
-
Genomic Diversity and Recombination Analysis of the Spike Protein Gene from Selected Human Coronaviruses.Biology (Basel). 2024 Apr 22;13(4):282. doi: 10.3390/biology13040282. Biology (Basel). 2024. PMID: 38666894 Free PMC article.
-
Ebselen and Diphenyl Diselenide Inhibit SARS-CoV-2 Replication at Non-Toxic Concentrations to Human Cell Lines.Vaccines (Basel). 2023 Jul 10;11(7):1222. doi: 10.3390/vaccines11071222. Vaccines (Basel). 2023. PMID: 37515038 Free PMC article.
References
-
- Ader F., Bouscambert-Duchamp M., Hites M., Peiffer-Smadja N., Poissy J., Belhadi D., Diallo A., Lê M.P., Peytavin G., Staub T., Greil R., Guedj J., Paiva J.A., Costagliola D., Yazdanpanah Y., Burdet C., Mentré F., Egle A., Greil R., Joannidis M., Lamprecht B., Altdorfer A., Belkhir L., Fraipont V., Hites M., Verschelden G., Aboab J., Ader F., Ait-Oufella H., Andrejak C., Andreu P., Argaud L., Bani-Sadr F., Benezit F., Blot M., Botelho-Nevers E., Bouadma L., Bouchaud O., Bougon D., Bouiller K., Bounes-Vardon F., Boutoille D., Boyer A., Bruel C., Cabié A., Canet E., Cazanave C., Chabartier C., Chirouze C., Clere-Jehl R., et al. Remdesivir plus standard of care versus standard of care alone for the treatment of patients admitted to hospital with COVID-19 (DisCoVeRy): a phase 3, randomised, controlled, open-label trial. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00485-0. - DOI - PMC - PubMed
-
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., MO Clarke, Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 - PMC - PubMed
-
- Alkhatib M., Svicher V., Salpini R., Ambrosio F.A., Bellocchi M.C., Carioti L., Piermatteo L., Scutari R., Costa G., Artese A., Alcaro S., Shafer R., Ceccherini-Silberstein F., Perez D.R. SARS-CoV-2 variants and their relevant mutational profiles: update summer 2021. Microbiol. Spectr. 2021;9 e01096-21. - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

