A RADAR method to measure DNA topoisomerase covalent complexes
- PMID: 35934484
- PMCID: PMC9722332
- DOI: 10.1016/bs.mie.2022.03.036
A RADAR method to measure DNA topoisomerase covalent complexes
Abstract
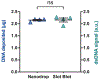
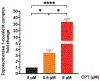
DNA topoisomerases resolve topological stress by introducing transient single- or double-strand breaks into the DNA duplex. This reaction requires the covalent binding of topoisomerases to DNA while the topological stress is being released. This transient intermediate is known as topoisomerase-covalent complex and represents the target of many anti-cancer drugs. Here, we describe a protocol to quantitatively detect topoisomerase-covalent complexes in vivo, called RADAR (rapid approach to DNA adduct recovery). DNA and protein-DNA covalent complexes are rapidly isolated from cells through chaotropic extraction. After normalization, samples are loaded on a slot blot, and the covalent complexes are detected using specific topoisomerase antibodies. In addition to being fast and robust, this assay produces quantitative results. Consequently, the RADAR assay can be applied to investigate the topoisomerase-covalent complex biology, including the effect of specific topoisomerase inhibitors. Finally, the same assay can be more generally applied to study covalent complexes of other enzymes with DNA.
Keywords: DNA topoisomerase; DNA topoisomerase–covalent complex; DNA-protein crosslink; Protein–DNA interaction; RADAR; Slot blot.
Copyright © 2022 Elsevier Inc. All rights reserved.
Figures

References
-
- Anand J, Sun Y, Zhao Y, Nitiss KC, & Nitiss JL (2018). Detection of topoisomerase covalent complexes in eukaryotic cells. In Drolet M (Ed.), DNA topoisomerases: Methods and protocols (pp. 283–299). New York, NY: Springer New York. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

