Phosphite synthetic auxotrophy as an effective biocontainment strategy for the industrial chassis Pseudomonas putida
- PMID: 35934698
- PMCID: PMC9358898
- DOI: 10.1186/s12934-022-01883-5
Phosphite synthetic auxotrophy as an effective biocontainment strategy for the industrial chassis Pseudomonas putida
Abstract
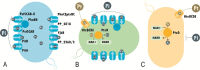
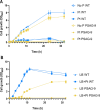
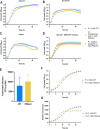
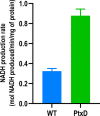
The inclusion of biosafety strategies into strain engineering pipelines is crucial for safe-by-design biobased processes. This in turn might enable a more rapid regulatory acceptance of bioengineered organisms in both industrial and environmental applications. For this reason, we equipped the industrially relevant microbial chassis Pseudomonas putida KT2440 with an effective biocontainment strategy based on a synthetic dependency on phosphite, which is generally not readily available in the environment. The produced PSAG-9 strain was first engineered to assimilate phosphite through the genome-integration of a phosphite dehydrogenase and a phosphite-specific transport complex. Subsequently, to deter the strain from growing on naturally assimilated phosphate, all native genes related to its transport were identified and deleted generating a strain unable to grow on media containing any phosphorous source other than phosphite. PSAG-9 exhibited fitness levels with phosphite similar to those of the wild type with phosphate, and low levels of escape frequency. Beyond biosafety, this strategy endowed P. putida with the capacity to be cultured under non-sterile conditions using phosphite as the sole phosphorous source with a reduced risk of contamination by other microbes, while displaying enhanced NADH regenerative capacity. These industrially beneficial features complement the metabolic advantages for which this species is known for, thereby strengthening it as a synthetic biology chassis with potential uses in industry, with suitability towards environmental release.
Keywords: Biocontainment; NADH; Non-sterile conditions; Phosphite; Pseudomonas putida; Synthetic auxotrophy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interest for this article.
Figures



References
-
- Fernández M, Duque E, Pizarro-Tobías P, Van Dillewijn P, Wittich R, Ramos JL. Microbial responses to xenobiotic compounds. Identification of genes that allow Pseudomonas putida KT2440 to cope with 2,4,6-trinitrotoluene. Microb Biotechnol. 2009;2:287–294. doi: 10.1111/j.1751-7915.2009.00085.x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

