ERCC5, HES6 and RORA are potential diagnostic markers of coronary artery disease
- PMID: 35934844
- PMCID: PMC9527589
- DOI: 10.1002/2211-5463.13469
ERCC5, HES6 and RORA are potential diagnostic markers of coronary artery disease
Abstract
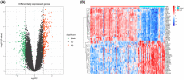
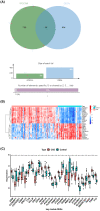
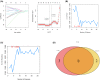
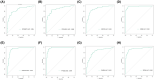
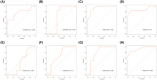
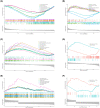
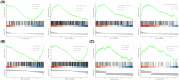
The mortality rate of patients with coronary artery disease (CAD) increases year by year, and the age of onset is decreasing, primarily because of the lack of an efficient and convenient diagnostic method for CAD. In the present study, we aimed to detect CAD-correlated biomarkers and the regulatory pathways involved through weighted co-expression network analysis. The microarray data originated from 93 CAD patients and 48 controls within the Gene Expression Omnibus (GEO) database. The gene network was implemented by weighted gene co-expression network analysis, and the genes were observed to fall into a range of modules. We took the intersection of genes in the modules most correlated with CAD with the differentially expressed genes of CAD, which were identified by applying the limma package. Lasso regression and support vector machine recursive feature elimination algorithms were used to determine CAD candidate signature genes. The biomarkers for diagnosing CAD were detected by validating candidate signature gene diagnostic capabilities (receiver operating characteristic curves) based on data sets from GEO. Three modules were selected, and 26 vital genes were identified. Eight of these genes were reported as the optimal candidate features in terms of CAD diagnosis. Through receiver operating characteristic curve analysis, we identified three genes (ERCC5, HES6 and RORA; area under the curve > 0.8) capable of distinguishing CAD from the control, and observed that these genes are correlated with the immune response. In summary, ERCC5, HES6 and RORA may have potential for diagnosis of CAD.
Keywords: ERCC5; HES6; RORA; bioinformatics; coronary heart disease; diagnostic marker.
© 2022 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Shao C, Wang J, Tian J, Tang Y‐D. Coronary artery disease: from mechanism to clinical practice. In: Wang M, editor. Coronary artery disease: therapeutics and drug discovery. Singapore: Springer Singapore; 2020. p. 1–36. - PubMed
-
- Shimada K. Immune system and atherosclerotic disease heterogeneity of leukocyte subsets participating in the pathogenesis of atherosclerosis. Circ Res. 2009;73:994–1001. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

