Prophage Diversity Across Salmonella and Verotoxin-Producing Escherichia coli in Agricultural Niches of British Columbia, Canada
- PMID: 35935192
- PMCID: PMC9355379
- DOI: 10.3389/fmicb.2022.853703
Prophage Diversity Across Salmonella and Verotoxin-Producing Escherichia coli in Agricultural Niches of British Columbia, Canada
Abstract
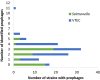
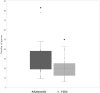
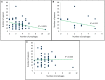
Prophages have long been regarded as an important contributor to the evolution of Salmonella and Verotoxin-producing E. coli (VTEC), members of the Enterobacteriaceae that cause millions of cases of foodborne illness in North America. In S. Typhimurium, prophages provide many of the genes required for invasion; similarly, in VTEC, the Verotoxin-encoding genes are located in cryptic prophages. The ability of prophages to quickly acquire and lose genes have driven their rapid evolution, leading to highly diversified populations of phages that can infect distantly-related bacterial hosts. To defend against foreign genetic materials (i.e., phages), bacteria have evolved Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) immunity, consisting of variable spacer regions that match short nucleic acid sequences of invaders previously encountered. The number of spacer regions varies widely amongst Enterobacteriaceae, and there is currently no clear consensus if the accumulation of spacers is linked to genomic prophage abundance. Given the immense prophage diversity and contribution to bacterial host phenotypes, we analyzed the prophage sequences within 118 strains of Salmonella and VTEC, 117 of which are of agricultural origin. Overall, 130 unique prophage sequences were identified and they were found to be remarkably diverse with <50% nucleotide similarity, particularly with the Gifsy-1 group which was identified in several Salmonella serovars and interestingly, a strain of VTEC. Additionally, we identified a novel plasmid-like phage that carried antibiotic resistance and bacteriocin resistance genes. The strains analyzed carried at least six distinct spacers which did not possess homology to prophages identified in the same genome. In fact, only a fraction of all identified spacers (14%) possessed significant homology to known prophages. Regression models did not discern a correlation between spacer and prophage abundance in our strains, although the relatively high number of spacers in our strains (an average of 27 in Salmonella and 19 in VTEC) suggest that high rates of infection may occur in agricultural niches and be a contributing driver in bacterial evolution. Cumulatively, these results shed insight into prophage diversity of Salmonella and VTEC, which will have further implications when informing development of phage therapies against these foodborne pathogens.
Keywords: CRISPR; Salmonella; VTEC; phage; prophage.
Copyright © 2022 Fong, Lu, Brenner, Falardeau and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Asadulghani M. D., Ogura Y., Ooka T., Itoh T., Sawaguchi A., Iguchi A., et al. (2009). The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5, e10000408. 10.1371/journal.ppat.1000408 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

