Isolation and characterization of two homolog phages infecting Pseudomonas aeruginosa
- PMID: 35935197
- PMCID: PMC9348578
- DOI: 10.3389/fmicb.2022.946251
Isolation and characterization of two homolog phages infecting Pseudomonas aeruginosa
Abstract
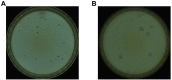
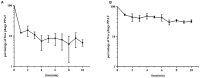
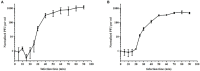
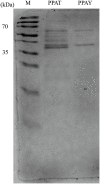
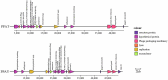
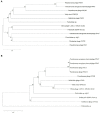
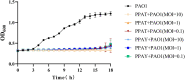
Bacteriophages (phages) are capable of infecting specific bacteria, and therefore can be used as a biological control agent to control bacteria-induced animal, plant, and human diseases. In this study, two homolog phages (named PPAY and PPAT) that infect Pseudomonas aeruginosa PAO1 were isolated and characterized. The results of the phage plaque assay showed that PPAT plaques were transparent dots, while the PPAY plaques were translucent dots with a halo. Transmission electron microscopy results showed that PPAT (65 nm) and PPAY (60 nm) strains are similar in size and have an icosahedral head and a short tail. Therefore, these belong to the short-tailed phage family Podoviridae. One-step growth curves revealed the latent period of 20 min and burst time of 30 min for PPAT and PPAY. The burst size of PPAT (953 PFUs/infected cell) was higher than that of PPAY (457 PFUs/infected cell). Also, the adsorption rate constant of PPAT (5.97 × 10-7 ml/min) was higher than that of PPAY (1.32 × 10-7 ml/min) at 5 min. Whole-genome sequencing of phages was carried out using the Illumina HiSeq platform. The genomes of PPAT and PPAY have 54,888 and 50,154 bp, respectively. Only 17 of the 352 predicted ORFs of PPAT could be matched to homologous genes of known function. Likewise, among the 351 predicted ORFs of PPAY, only 18 ORFs could be matched to genes of established functions. Homology and evolutionary analysis indicated that PPAT and PPAY are closely related to PA11. The presence of tail fiber proteins in PPAY but not in PPAT may have contributed to the halo effect of its plaque spots. In all, PPAT and PPAY, newly discovered P. aeruginosa phages, showed growth inhibitory effects on bacteria and can be used for research and clinical purposes.
Keywords: Pseudomonas aeruginosa; bacteriophages; genome; phage therapy; tail fiber protein.
Copyright © 2022 Yuanyuan, Xiaobo, Shang, Yutong, Hongrui, Chenyu, Bin, Xi, Chen, Zhiqiang, Jingfeng, Yun, Pingfeng and Zhigang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Isolation and characterization of T7-like lytic bacteriophages infecting multidrug resistant Pseudomonas aeruginosa isolated from Egypt.Curr Microbiol. 2015 Jun;70(6):786-91. doi: 10.1007/s00284-015-0788-8. Epub 2015 Feb 18. Curr Microbiol. 2015. PMID: 25691338
-
Isolation and Characterization of Lytic Bacteriophages Capable of Infecting Diverse Multidrug-Resistant Strains of Pseudomonas aeruginosa: PaCCP1 and PaCCP2.Pharmaceuticals (Basel). 2024 Nov 30;17(12):1616. doi: 10.3390/ph17121616. Pharmaceuticals (Basel). 2024. PMID: 39770458 Free PMC article.
-
Characterization and Genomic Analyses of Pseudomonas aeruginosa Podovirus TC6: Establishment of Genus Pa11virus.Front Microbiol. 2018 Oct 25;9:2561. doi: 10.3389/fmicb.2018.02561. eCollection 2018. Front Microbiol. 2018. PMID: 30410478 Free PMC article.
-
Characterization of Pseudomonas aeruginosa bacteriophages and control hemorrhagic pneumonia on a mice model.Front Microbiol. 2024 May 14;15:1396774. doi: 10.3389/fmicb.2024.1396774. eCollection 2024. Front Microbiol. 2024. PMID: 38808279 Free PMC article.
-
Genomic and Transcriptional Mapping of PaMx41, Archetype of a New Lineage of Bacteriophages Infecting Pseudomonas aeruginosa.Appl Environ Microbiol. 2016 Oct 27;82(22):6541-6547. doi: 10.1128/AEM.01415-16. Print 2016 Nov 15. Appl Environ Microbiol. 2016. PMID: 27590812 Free PMC article.
Cited by
-
Isolation and Characterization of a Lytic Phage PaTJ Against Pseudomonas aeruginosa.Viruses. 2024 Nov 21;16(12):1816. doi: 10.3390/v16121816. Viruses. 2024. PMID: 39772127 Free PMC article.
-
Isolation, characterization, and genomic analysis of a novel bacteriophage MA9V-1 infecting Chryseobacterium indologenes: a pathogen of Panax notoginseng root rot.Front Microbiol. 2023 Sep 14;14:1251211. doi: 10.3389/fmicb.2023.1251211. eCollection 2023. Front Microbiol. 2023. PMID: 37779709 Free PMC article.
-
A Novel Bacteriophage Infecting Multi-Drug- and Extended-Drug-Resistant Pseudomonas aeruginosa Strains.Antibiotics (Basel). 2024 Jun 3;13(6):523. doi: 10.3390/antibiotics13060523. Antibiotics (Basel). 2024. PMID: 38927189 Free PMC article.
-
Genomic characterization of a novel Pseudomonas aeruginosa bacteriophage representing the newly proposed genus Angoravirus: in vitro antimicrobial and antibiofilm activity.Int Microbiol. 2025 May 8. doi: 10.1007/s10123-025-00669-0. Online ahead of print. Int Microbiol. 2025. PMID: 40338461
References
-
- Ahamed S. T., Roy B., Basu U., Dutta S., Ghosh A. N., Bandyopadhyay B., et al. . (2019). Genomic and proteomic characterizations of Sfin-1, a novel lytic phage infecting multidrug-resistant Shigella spp. and Escherichia coli C. Front. Microbiol. 10:1876. doi: 10.3389/fmicb.2019.01876, PMID: - DOI - PMC - PubMed
-
- Alič Š., Naglič T., Tušek-Žnidarič M., Ravnikar M., Rački N., Peterka M., et al. . (2017). Newly isolated bacteriophages from the Podoviridae, Siphoviridae, and Myoviridae families have variable effects on putative novel Dickeya spp. Front. Microbiol. 8:1870. doi: 10.3389/fmicb.2017.01870, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

