Biomarker enrichment medium: A defined medium for metabolomic analysis of microbial pathogens
- PMID: 35935218
- PMCID: PMC9354526
- DOI: 10.3389/fmicb.2022.957158
Biomarker enrichment medium: A defined medium for metabolomic analysis of microbial pathogens
Abstract
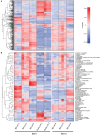
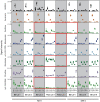
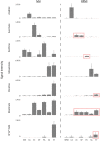
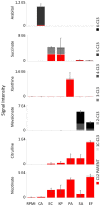
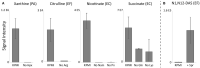
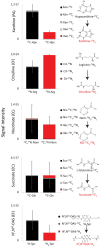
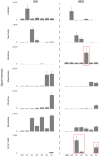
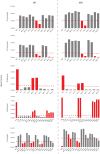
Microbes have diverse metabolic capabilities and differences in these phenotypes are critical for differentiating strains, species, and broader taxa of microorganisms. Recent advances in liquid chromatography-mass spectrometry (LC-MS) allow researchers to track the complex combinations of molecules that are taken up by each cell type and to quantify the rates that individual metabolites enter or exit the cells. This metabolomics-based approach allows complex metabolic phenotypes to be captured in a single assay, enables computational models of microbial metabolism to be constructed, and can serve as a diagnostic approach for clinical microbiology. Unfortunately, metabolic phenotypes are directly affected by the molecular composition of the culture medium and many traditional media are subject to molecular-level heterogeneity. Herein, we show that commercially sourced Mueller Hinton (MH) medium, a Clinical and Laboratory Standards Institute (CLSI) approved medium for clinical microbiology, has significant lot-to-lot and supplier-to-supplier variability in the concentrations of individual nutrients. We show that this variability does not affect microbial growth rates but does affect the metabolic phenotypes observed in vitro-including metabolic phenotypes that distinguish six common pathogens. To address this, we used a combination of isotope-labeling, substrate exclusion, and nutritional supplementation experiments using Roswell Park Memorial Institute (RPMI) medium to identify the specific nutrients used by the microbes to produce diagnostic biomarkers, and to formulate a Biomarker Enrichment Medium (BEM) as an alternative to complex undefined media for metabolomics research, clinical diagnostics, antibiotic susceptibility testing, and other applications where the analysis of stable microbial metabolic phenotypes is important.
Keywords: LC-MS; Mueller Hinton; biomarker enrichment medium; biomarkers; metabolomics.
Copyright © 2022 Mapar, Rydzak, Groves and Lewis.
Figures
References
-
- Clinical and Laboratory Standards Institute . (2022). M100: Performance Standards for Antimicrobial Susceptibility Testing, 32nd Edn. p. 1–362.
-
- Cocaign-Bousquet M., Garrigues C., Novak L., Lindley N., Loublere P. (1995). Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J. Appl. Bacteriol. 79, 108–116. 10.1111/j.1365-2672.1995.tb03131.x - DOI
LinkOut - more resources
Full Text Sources

