Phenotypic and integrated analysis of a comprehensive Pseudomonas aeruginosa PAO1 library of mutants lacking cyclic-di-GMP-related genes
- PMID: 35935233
- PMCID: PMC9355167
- DOI: 10.3389/fmicb.2022.949597
Phenotypic and integrated analysis of a comprehensive Pseudomonas aeruginosa PAO1 library of mutants lacking cyclic-di-GMP-related genes
Abstract
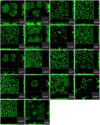
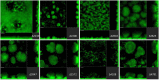
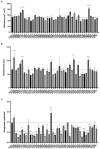
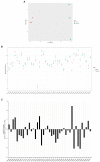
Pseudomonas aeruginosa is a Gram-negative bacterium that is able to survive and adapt in a multitude of niches as well as thrive within many different hosts. This versatility lies within its large genome of ca. 6 Mbp and a tight control in the expression of thousands of genes. Among the regulatory mechanisms widespread in bacteria, cyclic-di-GMP signaling is one which influences all levels of control. c-di-GMP is made by diguanylate cyclases and degraded by phosphodiesterases, while the intracellular level of this molecule drives phenotypic responses. Signaling involves the modification of enzymes' or proteins' function upon c-di-GMP binding, including modifying the activity of regulators which in turn will impact the transcriptome. In P. aeruginosa, there are ca. 40 genes encoding putative DGCs or PDEs. The combined activity of those enzymes should reflect the overall c-di-GMP concentration, while specific phenotypic outputs could be correlated to a given set of dgc/pde. This notion of specificity has been addressed in several studies and different strains of P. aeruginosa. Here, we engineered a mutant library for the 41 individual dgc/pde genes in P. aeruginosa PAO1. In most cases, we observed a significant to slight variation in the global c-di-GMP pool of cells grown planktonically, while several mutants display a phenotypic impact on biofilm including initial attachment and maturation. If this observation of minor changes in c-di-GMP level correlating with significant phenotypic impact appears to be true, it further supports the idea of a local vs global c-di-GMP pool. In contrast, there was little to no effect on motility, which differs from previous studies. Our RNA-seq analysis indicated that all PAO1 dgc/pde genes were expressed in both planktonic and biofilm growth conditions and our work suggests that c-di-GMP networks need to be reconstructed for each strain separately and cannot be extrapolated from one to another.
Keywords: Pseudomonas; biofilm; c-di-GMP; diguanylate cyclase; phosphodiesterase.
Copyright © 2022 Eilers, Kuok Hoong Yam, Morton, Mei Hui Yong, Brizuela, Hadjicharalambous, Liu, Givskov, Rice and Filloux.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3'-5')-cyclic-GMP in virulence.Proc Natl Acad Sci U S A. 2006 Feb 21;103(8):2839-44. doi: 10.1073/pnas.0511090103. Epub 2006 Feb 13. Proc Natl Acad Sci U S A. 2006. PMID: 16477007 Free PMC article.
-
FlhF affects the subcellular clustering of WspR through HsbR in Pseudomonas aeruginosa.Appl Environ Microbiol. 2024 Jan 24;90(1):e0154823. doi: 10.1128/aem.01548-23. Epub 2023 Dec 19. Appl Environ Microbiol. 2024. PMID: 38112425 Free PMC article.
-
ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa.PLoS Pathog. 2014 Mar 6;10(3):e1003984. doi: 10.1371/journal.ppat.1003984. eCollection 2014 Mar. PLoS Pathog. 2014. PMID: 24603766 Free PMC article.
-
c-di-GMP signaling in Pseudomonas syringae complex.Microbiol Res. 2023 Oct;275:127445. doi: 10.1016/j.micres.2023.127445. Epub 2023 Jul 6. Microbiol Res. 2023. PMID: 37450986 Review.
-
Molecular and structural facets of c-di-GMP signalling associated with biofilm formation in Pseudomonas aeruginosa.Mol Aspects Med. 2021 Oct;81:101001. doi: 10.1016/j.mam.2021.101001. Epub 2021 Jul 24. Mol Aspects Med. 2021. PMID: 34311995 Review.
Cited by
-
Control of biofilm formation by an Agrobacterium tumefaciens pterin-binding periplasmic protein conserved among diverse Proteobacteria.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2319903121. doi: 10.1073/pnas.2319903121. Epub 2024 Jun 13. Proc Natl Acad Sci U S A. 2024. PMID: 38870058 Free PMC article.
-
Genetic Dissection of Cyclic di-GMP Signalling in Pseudomonas aeruginosa via Systematic Diguanylate Cyclase Disruption.Microb Biotechnol. 2025 Apr;18(4):e70137. doi: 10.1111/1751-7915.70137. Microb Biotechnol. 2025. PMID: 40172309 Free PMC article.
-
Echinacoside reduces intracellular c-di-GMP levels and potentiates tobramycin activity against Pseudomonas aeruginosa biofilm aggregates.NPJ Biofilms Microbiomes. 2025 Mar 7;11(1):40. doi: 10.1038/s41522-025-00673-2. NPJ Biofilms Microbiomes. 2025. PMID: 40055321 Free PMC article.
-
Control of Biofilm Formation by an Agrobacterium tumefaciens Pterin-Binding Periplasmic Protein Conserved Among Pathogenic Bacteria.bioRxiv [Preprint]. 2023 Nov 21:2023.11.18.567607. doi: 10.1101/2023.11.18.567607. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2319903121. doi: 10.1073/pnas.2319903121. PMID: 38014264 Free PMC article. Updated. Preprint.
-
Student-led experimental evolution reveals novel biofilm regulators of adaptation to multiple niches.bioRxiv [Preprint]. 2025 Jul 1:2025.06.06.658356. doi: 10.1101/2025.06.06.658356. bioRxiv. 2025. PMID: 40501954 Free PMC article. Preprint.
References
-
- Andersen J. B., Hultqvist L. D., Jansen C. U., Jakobsen T. H., Nilsson M., Rybtke M., et al. . (2021). Identification of small molecules that interfere with c-di-GMP signaling and induce dispersal of Pseudomonas aeruginosa biofilms. NPJ Biofilms Microbiomes 7:59. doi: 10.1038/s41522-021-00225-4, PMID: - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

