Combining Metabolic Engineering and Multiplexed Screening Methods for 3-Hydroxypropionic Acid Production in Pichia pastoris
- PMID: 35935509
- PMCID: PMC9354023
- DOI: 10.3389/fbioe.2022.942304
Combining Metabolic Engineering and Multiplexed Screening Methods for 3-Hydroxypropionic Acid Production in Pichia pastoris
Erratum in
-
Corrigendum: Combining metabolic engineering and multiplexed screening methods for 3-hydroxypropionic acid production in Pichia pastoris.Front Bioeng Biotechnol. 2022 Sep 30;10:1003012. doi: 10.3389/fbioe.2022.1003012. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36246370 Free PMC article.
Abstract
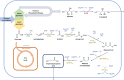
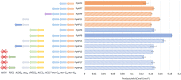
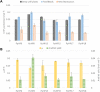
Production of 3-hydroxypropionic acid (3-HP) in Pichia pastoris (syn. Komagataella phaffii) via the malonyl-CoA pathway has been recently demonstrated using glycerol as a carbon source, but the reported metrics were not commercially relevant. The flux through the heterologous pathway from malonyl-CoA to 3-HP was hypothesized as the main bottleneck. In the present study, different metabolic engineering approaches have been combined to improve the productivity of the original 3-HP producing strains. To do so, an additional copy of the gene encoding for the potential rate-limiting step of the pathway, i.e., the C-terminal domain of the malonyl-CoA reductase, was introduced. In addition, a variant of the endogenous acetyl-CoA carboxylase (ACC1 S1132A ) was overexpressed with the aim to increase the delivery of malonyl-CoA. Furthermore, the genes encoding for the pyruvate decarboxylase, aldehyde dehydrogenase and acetyl-CoA synthase, respectively, were overexpressed to enhance conversion of pyruvate into cytosolic acetyl-CoA, and the main gene responsible for the production of the by-product D-arabitol was deleted. Three different screening conditions were used to classify the performance of the different strains: 24-deep-well plates batch cultures, small-scale cultures in falcon tubes using FeedBeads® (i.e., slow release of glycerol over time), and mini bioreactor batch cultures. The best two strains from the FeedBeads® screening, PpHP8 and PpHP18, were tested in bioreactor fed-batch cultures using a pre-fixed exponentially increasing feeding rate. The strain PpHP18 produced up to 37.05 g L-1 of 3-HP at 0.712 g L-1 h-1 with a final product yield on glycerol of 0.194 Cmol-1 in fed-batch cultures. Remarkably, PpHP18 did not rank among the 2-top producer strains in small scale batch cultivations in deep-well plates and mini bioreactors, highlighting the importance of multiplexed screening conditions for adequate assessment of metabolic engineering strategies. These results represent a 50% increase in the product yield and final concentration, as well as over 30% increase in volumetric productivity compared to the previously obtained metrics for P. pastoris. Overall, the combination of glycerol as carbon source and a metabolically engineered P. pastoris strain resulted in the highest 3-HP concentration and productivity reported so far in yeast.
Keywords: 3-hydroxypropionic acid; Pichia pastoris; acetyl-CoA; glycerol; malonyl-CoA; metabolic engineering.
Copyright © 2022 Fina, Heux, Albiol and Ferrer.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Chemarin F., Athès V., Bedu M., Loty T., Allais F., Trelea I. C., et al. (2019). Towards an In Situ Product Recovery of Bio-Based 3-hydroxypropionic Acid: Influence of Bioconversion Broth Components on Membrane-Assisted Reactive Extraction. J. Chem. Technol. Biotechnol. 94, 964–972. 10.1002/jctb.5845 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

