A numerical population density technique for N-dimensional neuron models
- PMID: 35935536
- PMCID: PMC9354936
- DOI: 10.3389/fninf.2022.883796
A numerical population density technique for N-dimensional neuron models
Abstract
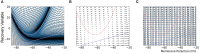
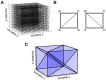
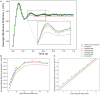
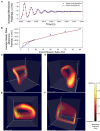
Population density techniques can be used to simulate the behavior of a population of neurons which adhere to a common underlying neuron model. They have previously been used for analyzing models of orientation tuning and decision making tasks. They produce a fully deterministic solution to neural simulations which often involve a non-deterministic or noise component. Until now, numerical population density techniques have been limited to only one- and two-dimensional models. For the first time, we demonstrate a method to take an N-dimensional underlying neuron model and simulate the behavior of a population. The technique enables so-called graceful degradation of the dynamics allowing a balance between accuracy and simulation speed while maintaining important behavioral features such as rate curves and bifurcations. It is an extension of the numerical population density technique implemented in the MIIND software framework that simulates networks of populations of neurons. Here, we describe the extension to N dimensions and simulate populations of leaky integrate-and-fire neurons with excitatory and inhibitory synaptic conductances then demonstrate the effect of degrading the accuracy on the solution. We also simulate two separate populations in an E-I configuration to demonstrate the technique's ability to capture complex behaviors of interacting populations. Finally, we simulate a population of four-dimensional Hodgkin-Huxley neurons under the influence of noise. Though the MIIND software has been used only for neural modeling up to this point, the technique can be used to simulate the behavior of a population of agents adhering to any system of ordinary differential equations under the influence of shot noise. MIIND has been modified to render a visualization of any three of an N-dimensional state space of a population which encourages fast model prototyping and debugging and could prove a useful educational tool for understanding dynamical systems.
Keywords: Python; dynamical systems; network; neural population; population density; simulator; software; visualization.
Copyright © 2022 Osborne and de Kamps.
Figures







References
LinkOut - more resources
Full Text Sources

