Draft genome sequences of hydrocarbon degrading Haloferax sp. AB510, Haladaptatus sp. AB618 and Haladaptatus sp. AB643 isolated from the estuarine sediments of Sundarban mangrove forests, India
- PMID: 35935548
- PMCID: PMC9349328
- DOI: 10.1007/s13205-022-03273-5
Draft genome sequences of hydrocarbon degrading Haloferax sp. AB510, Haladaptatus sp. AB618 and Haladaptatus sp. AB643 isolated from the estuarine sediments of Sundarban mangrove forests, India
Abstract
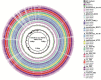
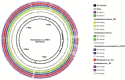
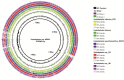
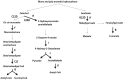
The present study reports the draft genomes of three hydrocarbon-degrading haloarchaeal strains Haloferax sp. AB510, Haladaptatus sp. AB618 and Haladaptatus sp. AB643 that were isolated from the estuarine sediments of Sundarban mangrove forests, India. All three genomes had a high GC content of around 60%, characteristic of the haloarchaea. The Haloferax sp. AB510 genome was around 3.9 Mb in size and consisted of 4567 coding sequences and 54 RNAs. The Haladaptatus sp. AB618 and Haladaptatus sp. AB643 genomes were comparatively larger and around 4.8 Mb each. The AB618 and AB643 genomes comprised 5279 and 5304 coding sequences and 60 and 59 RNAs, respectively. All three of the genomes encoded several genes that attributed to their survival in the presence of hydrocarbons in their native habitats. Functional annotation and curation of the sequenced genomes suggested that the Haloferax sp. AB510 strain utilized the gentisate pathway of aromatic compound degradation. While the Haladaptatus sp. AB618 and Haladaptatus sp. AB643 strains possessed the freedom of utilizing both the gentisate and the catechol pathways for degrading aromatic hydrocarbons.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-022-03273-5.
© King Abdulaziz City for Science and Technology 2022.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures

References
-
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. - DOI - PMC - PubMed
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

