Identification of Genes Predicting Poor Response of Trastuzumab in Human Epidermal Growth Factor Receptor 2 Positive Breast Cancer
- PMID: 35935587
- PMCID: PMC9348965
- DOI: 10.1155/2022/9529114
Identification of Genes Predicting Poor Response of Trastuzumab in Human Epidermal Growth Factor Receptor 2 Positive Breast Cancer
Abstract
Objective: To identify trastuzumab-resistant genes predicting drug response and poor prognosis in human epidermal growth factor receptor 2 positive (HER2+) breast cancer.
Methods: Gene expression profiles from the GEO (Gene Expression Omnibus) database were obtained and analyzed. Differentially expressed genes (DEGs) between the pathological complete response (pCR) group and non-pCR group in a trastuzumab neoadjuvant therapy cohort and DEGs between Herceptin-resistant and wild-type cell lines were detected and evaluated. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analyses were performed to select the functional hub genes. The hub genes' prognostic power was validated by another trastuzumab adjuvant treatment cohort.
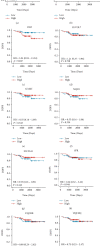
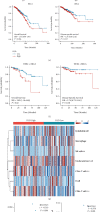
Results: Fifty upregulated overlapping DEGs were identified by analyzing two trastuzumab resistance-related GEO databases. Functional analysis picked out ten hub genes enriched in mitochondrial function and metabolism pathways: ASCL1, CPT2, DLD, ELVOL7, GAMT, NQO1, SLC23A1, SPR, UQCRB, and UQCRQ. These hub genes could distinguish patients with trastuzumab resistance from the sensitive ones. Further survival analysis of hub genes showed that DLD overexpression was significantly associated with an unfavorable prognosis in HER2+ breast cancer patients.
Conclusion: Ten novel trastuzumab resistance-related genes were discovered, of which DLD could be used for trastuzumab response prediction and prognostic prediction in HER2+ breast cancer.
Copyright © 2022 Xinrui Dong et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Hayashi N., Niikura N., Yamauchi H., Nakamura S., Ueno N. T. Adding hormonal therapy to chemotherapy and trastuzumab improves prognosis in patients with hormone receptor-positive and human epidermal growth factor receptor 2-positive primary breast cancer. Breast Cancer Research and Treatment . 2013;137(2, article 2336):523–531. doi: 10.1007/s10549-012-2336-6. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

