Sex-dependent transcription of cardiac electrophysiology and links to acetylation modifiers based on the GTEx database
- PMID: 35935618
- PMCID: PMC9354462
- DOI: 10.3389/fcvm.2022.941890
Sex-dependent transcription of cardiac electrophysiology and links to acetylation modifiers based on the GTEx database
Abstract
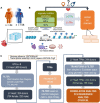
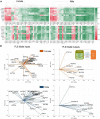
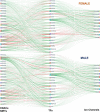
Development of safer drugs based on epigenetic modifiers, e.g., histone deacetylase inhibitors (HDACi), requires better understanding of their effects on cardiac electrophysiology. Using RNAseq data from the genotype-tissue-expression database (GTEx), we created models that link the abundance of acetylation enzymes (HDAC/SIRT/HATs), and the gene expression of ion channels (IC) via select cardiac transcription factors (TFs) in male and female adult human hearts (left ventricle, LV). Gene expression data (transcripts per million, TPM) from GTEx donors (21-70 y.o.) were filtered, normalized and transformed to Euclidian space to allow quantitative comparisons in 84 female and 158 male LVs. Sex-specific partial least-square (PLS) regression models, linking gene expression data for HDAC/SIRT/HATs to TFs and to ICs gene expression, revealed tight co-regulation of cardiac ion channels by HDAC/SIRT/HATs, with stronger clustering in the male LV. Co-regulation of genes encoding excitatory and inhibitory processes in cardiac tissue by the acetylation modifiers may help explain their predominantly net-neutral effects on cardiac electrophysiology. ATP1A1, encoding for the Na/K pump, represented an outlier-with orthogonal regulation by the acetylation modifiers to most of the ICs. The HDAC/SIRT/HAT effects were mediated by strong (+) TF regulators of ICs, e.g., MEF2A and TBX5, in both sexes. Furthermore, for male hearts, PLS models revealed a stronger (+/-) mediatory role on ICs for NKX25 and TGF1B/KLF4, respectively, while RUNX1 exhibited larger (-) TF effects on ICs in females. Male-trained PLS models of HDAC/SIRT/HAT effects on ICs underestimated the effects on some ICs in females. Insights from the GTEx dataset about the co-expression and transcriptional co-regulation of acetylation-modifying enzymes, transcription factors and key cardiac ion channels in a sex-specific manner can help inform safer drug design.
Keywords: GTEx; PLS; cardiac electrophysiology; epigenetics; histone acetylation; left ventricle; sex differences.
Copyright © 2022 Pressler, Horvath and Entcheva.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
HDAC inhibition attenuates cardiac hypertrophy by acetylation and deacetylation of target genes.Epigenetics. 2015;10(5):418-30. doi: 10.1080/15592294.2015.1024406. Epub 2015 May 5. Epigenetics. 2015. PMID: 25941940 Free PMC article.
-
Pathological histone acetylation in Parkinson's disease: Neuroprotection and inhibition of microglial activation through SIRT 2 inhibition.Neurosci Lett. 2018 Feb 14;666:48-57. doi: 10.1016/j.neulet.2017.12.037. Epub 2017 Dec 19. Neurosci Lett. 2018. PMID: 29273397 Free PMC article.
-
Mechanism of histone deacetylases in cardiac hypertrophy and its therapeutic inhibitors.Front Cardiovasc Med. 2022 Jul 26;9:931475. doi: 10.3389/fcvm.2022.931475. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35958418 Free PMC article. Review.
-
Imbalance between HAT and HDAC activities in the PBMCs of patients with ankylosing spondylitis or rheumatoid arthritis and influence of HDAC inhibitors on TNF alpha production.PLoS One. 2013 Aug 15;8(8):e70939. doi: 10.1371/journal.pone.0070939. eCollection 2013. PLoS One. 2013. PMID: 24039666 Free PMC article.
-
[Epigenetic mechanisms and alcohol use disorders: a potential therapeutic target].Biol Aujourdhui. 2017;211(1):83-91. doi: 10.1051/jbio/2017014. Epub 2017 Jul 6. Biol Aujourdhui. 2017. PMID: 28682229 Review. French.
Cited by
-
Non parametric differential network analysis: a tool for unveiling specific molecular signatures.BMC Bioinformatics. 2024 Nov 18;25(1):359. doi: 10.1186/s12859-024-05969-2. BMC Bioinformatics. 2024. PMID: 39558195 Free PMC article.
-
OptoDyCE-plate as an affordable high throughput imager for all optical cardiac electrophysiology.bioRxiv [Preprint]. 2023 Aug 31:2023.08.29.555447. doi: 10.1101/2023.08.29.555447. bioRxiv. 2023. Update in: J Mol Cell Cardiol Plus. 2023 Dec;6:100054. doi: 10.1016/j.jmccpl.2023.100054. PMID: 37693544 Free PMC article. Updated. Preprint.
-
OptoDyCE-plate as an affordable high throughput imager for all optical cardiac electrophysiology.J Mol Cell Cardiol Plus. 2023 Dec;6:100054. doi: 10.1016/j.jmccpl.2023.100054. Epub 2023 Nov 9. J Mol Cell Cardiol Plus. 2023. PMID: 38130942 Free PMC article.
-
High-throughput optical sensing of peri-cellular oxygen in cardiac cells: system characterization, calibration, and testing.bioRxiv [Preprint]. 2023 Apr 28:2023.04.24.538133. doi: 10.1101/2023.04.24.538133. bioRxiv. 2023. Update in: Front Bioeng Biotechnol. 2023 Jun 16;11:1214493. doi: 10.3389/fbioe.2023.1214493. PMID: 37163022 Free PMC article. Updated. Preprint.
-
Analysis of age-dependent gene-expression in human tissues for studying diabetes comorbidities.Sci Rep. 2023 Jun 26;13(1):10372. doi: 10.1038/s41598-023-37550-x. Sci Rep. 2023. PMID: 37365269 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

