Development and validation of a hypoxia-stemness-based prognostic signature in pancreatic adenocarcinoma
- PMID: 35935823
- PMCID: PMC9350896
- DOI: 10.3389/fphar.2022.939542
Development and validation of a hypoxia-stemness-based prognostic signature in pancreatic adenocarcinoma
Abstract
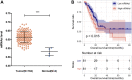
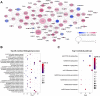
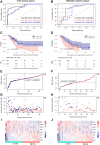
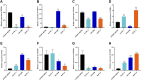
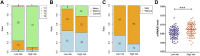
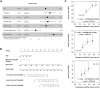
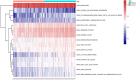
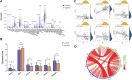
Background: Pancreatic adenocarcinoma (PAAD) is one of the most aggressive and fatal gastrointestinal malignancies with high morbidity and mortality worldwide. Accumulating evidence has revealed the clinical significance of the interaction between the hypoxic microenvironment and cancer stemness in pancreatic cancer progression and therapies. This study aims to identify a hypoxia-stemness index-related gene signature for risk stratification and prognosis prediction in PAAD. Methods: The mRNA expression-based stemness index (mRNAsi) data of PAAD samples from The Cancer Genome Atlas (TCGA) database were calculated based on the one-class logistic regression (OCLR) machine learning algorithm. Univariate Cox regression and LASSO regression analyses were then performed to establish a hypoxia-mRNAsi-related gene signature, and its prognostic performance was verified in both the TCGA-PAAD and GSE62452 corhorts by Kaplan-Meier and receiver operating characteristic (ROC) analyses. Additionally, we further validated the expression levels of signature genes using the TCGA, GTEx and HPA databases as well as qPCR experiments. Moreover, we constructed a prognostic nomogram incorporating the eight-gene signature and traditional clinical factors and analyzed the correlations of the risk score with immune infiltrates and immune checkpoint genes. Results: The mRNAsi values of PAAD samples were significantly higher than those of normal samples (p < 0.001), and PAAD patients with high mRNAsi values exhibited worse overall survival (OS). A novel prognostic risk model was successfully constructed based on the eight-gene signature comprising JMJD6, NDST1, ENO3, LDHA, TES, ANKZF1, CITED, and SIAH2, which could accurately predict the 1-, 3-, and 5-year OS of PAAD patients in both the training and external validation datasets. Additionally, the eight-gene signature could distinguish PAAD samples from normal samples and stratify PAAD patients into low- and high-risk groups with distinct OS. The risk score was closely correlated with immune cell infiltration patterns and immune checkpoint molecules. Moreover, calibration analysis showed the excellent predictive ability of the nomogram incorporating the eight-gene signature and traditional clinical factors. Conclusion: We developed a hypoxia-stemness-related prognostic signature that reliably predicts the OS of PAAD. Our findings may aid in the risk stratification and individual treatment of PAAD patients.
Keywords: cancer stemness; hypoxia microenvironment; mRNAsi; pancreatic adenocarcinoma; prognosis.
Copyright © 2022 Tian, Zheng, Mou, Lu, Chen, Du, Zheng, Chen, Shen, Li and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abou Khouzam R., Rao S. P., Venkatesh G. H., Zeinelabdin N. A., Buart S., Meylan M., et al. (2021). An eight-gene hypoxia signature predicts survival in pancreatic cancer and is associated with an immunosuppressed tumor microenvironment. Front. Immunol. 12, 680435. 10.3389/fimmu.2021.680435 - DOI - PMC - PubMed
-
- Aglietta M., Barone C., Sawyer M. B., Moore M. J., Miller W. H., Bagala C., et al. (2014). A phase I dose escalation trial of tremelimumab (CP-675, 206) in combination with gemcitabine in chemotherapy-naive patients with metastatic pancreatic cancer. Ann. Oncol. 25 (9), 1750–1755. 10.1093/annonc/mdu205 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

