Fungal Planet description sheets: 1182-1283
- PMID: 35935893
- PMCID: PMC9311394
- DOI: 10.3767/persoonia.2021.46.11
Fungal Planet description sheets: 1182-1283
Abstract
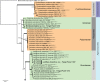
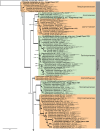
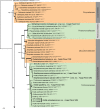
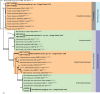
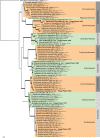
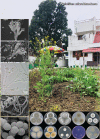
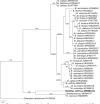
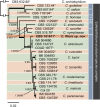
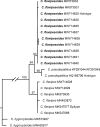
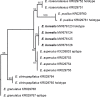
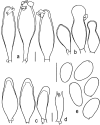
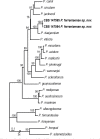
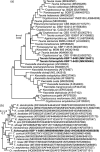
Novel species of fungi described in this study include those from various countries as follows: Algeria, Phaeoacremonium adelophialidum from Vitis vinifera. Antarctica, Comoclathris antarctica from soil. Australia, Coniochaeta salicifolia as endophyte from healthy leaves of Geijera salicifolia, Eremothecium peggii in fruit of Citrus australis, Microdochium ratticaudae from stem of Sporobolus natalensis, Neocelosporium corymbiae on stems of Corymbia variegata, Phytophthora kelmanii from rhizosphere soil of Ptilotus pyramidatus, Pseudosydowia backhousiae on living leaves of Backhousia citriodora, Pseudosydowia indooroopillyensis, Pseudosydowia louisecottisiae and Pseudosydowia queenslandica on living leaves of Eucalyptus sp. Brazil, Absidia montepascoalis from soil. Chile, Ilyonectria zarorii from soil under Maytenus boaria. Costa Rica, Colletotrichum filicis from an unidentified fern. Croatia, Mollisia endogranulata on deteriorated hardwood. Czech Republic, Arcopilus navicularis from tea bag with fruit tea, Neosetophoma buxi as endophyte from Buxus sempervirens, Xerochrysium bohemicum on surface of biscuits with chocolate glaze and filled with jam. France, Entoloma cyaneobasale on basic to calcareous soil, Fusarium aconidiale from Triticum aestivum, Fusarium juglandicola from buds of Juglans regia. Germany, Tetraploa endophytica as endophyte from Microthlaspi perfoliatum roots. India, Castanediella ambae on leaves of Mangifera indica, Lactifluus kanadii on soil under Castanopsis sp., Penicillium uttarakhandense from soil. Italy, Penicillium ferraniaense from compost. Namibia, Bezerromyces gobabebensis on leaves of unidentified succulent, Cladosporium stipagrostidicola on leaves of Stipagrostis sp., Cymostachys euphorbiae on leaves of Euphorbia sp., Deniquelata hypolithi from hypolith under a rock, Hysterobrevium walvisbayicola on leaves of unidentified tree, Knufia hypolithi and Knufia walvisbayicola from hypolith under a rock, Lapidomyces stipagrostidicola on leaves of Stipagrostis sp., Nothophaeotheca mirabibensis (incl. Nothophaeotheca gen. nov.) on persistent inflorescence remains of Blepharis obmitrata, Paramyrothecium salvadorae on twigs of Salvadora persica, Preussia procaviicola on dung of Procavia sp., Sordaria equicola on zebra dung, Volutella salvadorae on stems of Salvadora persica. Netherlands, Entoloma ammophilum on sandy soil, Entoloma pseudocruentatum on nutrient poor (acid) soil, Entoloma pudens on plant debris, amongst grasses. New Zealand, Amorocoelophoma neoregeliae from leaf spots of Neoregelia sp., Aquilomyces metrosideri and Septoriella callistemonis from stem discolouration and leaf spots of Metrosideros sp., Cadophora neoregeliae from leaf spots of Neoregelia sp., Flexuomyces asteliae (incl. Flexuomyces gen. nov.) and Mollisia asteliae from leaf spots of Astelia chathamica, Ophioceras freycinetiae from leaf spots of Freycinetia banksii, Phaeosphaeria caricis-sectae from leaf spots of Carex secta. Norway, Cuphophyllus flavipesoides on soil in semi-natural grassland, Entoloma coracis on soil in calcareous Pinus and Tilia forests, Entoloma cyaneolilacinum on soil semi-natural grasslands, Inocybe norvegica on gravelly soil. Pakistan, Butyriboletus parachinarensis on soil in association with Quercus baloot. Poland, Hyalodendriella bialowiezensis on debris beneath fallen bark of Norway spruce Picea abies. Russia, Bolbitius sibiricus on à moss covered rotting trunk of Populus tremula, Crepidotus wasseri on debris of Populus tremula, Entoloma isborscanum on soil on calcareous grasslands, Entoloma subcoracis on soil in subalpine grasslands, Hydropus lecythiocystis on rotted wood of Betula pendula, Meruliopsis faginea on fallen dead branches of Fagus orientalis, Metschnikowia taurica from fruits of Ziziphus jujube, Suillus praetermissus on soil, Teunia lichenophila as endophyte from Cladonia rangiferina. Slovakia, Hygrocybe fulgens on mowed grassland, Pleuroflammula pannonica from corticated branches of Quercus sp. South Africa, Acrodontium burrowsianum on leaves of unidentified Poaceae, Castanediella senegaliae on dead pods of Senegalia ataxacantha, Cladophialophora behniae on leaves of Behnia sp., Colletotrichum cliviigenum on leaves of Clivia sp., Diatrype dalbergiae on bark of Dalbergia armata, Falcocladium heteropyxidicola on leaves of Heteropyxis canescens, Lapidomyces aloidendricola as epiphyte on brown stem of Aloidendron dichotomum, Lasionectria sansevieriae and Phaeosphaeriopsis sansevieriae on leaves of Sansevieria hyacinthoides, Lylea dalbergiae on Diatrype dalbergiae on bark of Dalbergia armata, Neochaetothyrina syzygii (incl. Neochaetothyrina gen. nov.) on leaves of Syzygium chordatum, Nothophaeomoniella ekebergiae (incl. Nothophaeomoniella gen. nov.) on leaves of Ekebergia pterophylla, Paracymostachys euphorbiae (incl. Paracymostachys gen. nov.) on leaf litter of Euphorbia ingens, Paramycosphaerella pterocarpi on leaves of Pterocarpus angolensis, Paramycosphaerella syzygii on leaf litter of Syzygium chordatum, Parateichospora phoenicicola (incl. Parateichospora gen. nov.) on leaves of Phoenix reclinata, Seiridium syzygii on twigs of Syzygium chordatum, Setophoma syzygii on leaves of Syzygium sp., Starmerella xylocopis from larval feed of an Afrotropical bee Xylocopa caffra, Teratosphaeria combreti on leaf litter of Combretum kraussii, Teratosphaericola leucadendri on leaves of Leucadendron sp., Toxicocladosporium pterocarpi on pods of Pterocarpus angolensis. Spain, Cortinarius bonachei with Quercus ilex in calcareus soils, Cortinarius brunneovolvatus under Quercus ilex subsp. ballota in calcareous soil, Extremopsis radicicola (incl. Extremopsis gen. nov.) from root-associated soil in a wet heathland, Russula quintanensis on acidic soils, Tubaria vulcanica on volcanic lapilii material, Tuber zambonelliae in calcareus soil. Sweden, Elaphomyces borealis on soil under Pinus sylvestris and Betula pubescens. Tanzania, Curvularia tanzanica on inflorescence of Cyperus aromaticus. Thailand, Simplicillium niveum on Ophiocordyceps camponoti-leonardi on underside of unidentified dicotyledonous leaf. USA, Calonectria californiensis on leaves of Umbellularia californica, Exophiala spartinae from surface sterilised roots of Spartina alterniflora, Neophaeococcomyces oklahomaensis from outside wall of alcohol distillery. Vietnam, Fistulinella aurantioflava on soil. Morphological and culture characteristics are supported by DNA barcodes. Citation: Crous PW, Cowan DA, Maggs-Kölling, et al. 2021. Fungal Planet description sheets: 1182-1283. Persoonia 46: 313-528. https://doi.org/10.3767/persoonia.2021.46.11.
Keywords: ITS nrDNA barcodes; LSU; new taxa; systematics.
© 2021 Naturalis Biodiversity Center & Westerdijk Fungal Biodiversity Institute.
Figures




























































































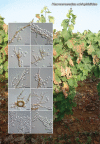


































References
-
- Abad ZG, Abad JA, Creswell T. 2002. Advances in the integration of morphological and molecular chraraterization in Phytophthora genus: The case of P. kelmania and other putative new species. Phytopathology 92 (6 suppl): S1.
-
- Abad ZG, Burgess T, Bienapfl JC, et al. 2019. IDphy: Molecular and morphological identification of Phytophthora based on the types. USDA APHIS PPQ S&T Beltsville Lab, USDA APHIS PPQ S&T ITP, Centre for Phytophthora Science and Management, and World Phytophthora Collection. https://idtools.org/id/phytophthora/index.php <different dates in 2020>.
-
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723.
-
- Alcorn JL. 1982. Ovariicolous bipolaris species on Sporobolus and other grasses. Mycotaxon 15: 20–48.
LinkOut - more resources
Full Text Sources
