Diversity of Backusella (Mucoromycotina) in south-eastern Australia revealed through polyphasic taxonomy
- PMID: 35935894
- PMCID: PMC9311390
- DOI: 10.3767/persoonia.2021.46.01
Diversity of Backusella (Mucoromycotina) in south-eastern Australia revealed through polyphasic taxonomy
Abstract
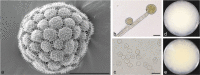
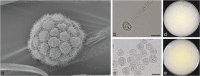
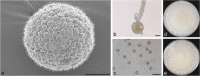
Here we explore the diversity of one morphologically distinguishable genus in the Mucoromycotina, Backusella, in south-eastern Australia. We isolated more than 200 strains from locations across the states of Victoria and Tasmania. Characterization of these strains using a combination of approaches including morphology, sucrose utilization and whole genome sequencing for 13 strains, revealed 10 new species. The genetic basis for interspecies variation in sucrose utilization was found to be the presence of a gene encoding an invertase enzyme. The genus Backusella is revised and a new key for species identification produced. Given that we have more than doubled the number of species in this genus, this work demonstrates that there may be considerable undiscovered species diversity in the early diverging fungal lineages. Citation: Urquhart AS, Douch JK, Heafield TA, et al. 2021. Diversity of Backusella (Mucoromycotina) in south-eastern Australia revealed through polyphasic taxonomy. Persoonia 46: 1-25. https://doi.org/10.3767/persoonia.2021.46.01.
Keywords: Backusella; Mucorales; genome sequencing; invertase; new taxa; polyphasic taxonomy; zygospore.
© 2020-2021 Naturalis Biodiversity Center & Westerdijk Fungal Biodiversity Institute.
Figures






















References
-
- Altschul SF, Gish W, Miller W, et al. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. - PubMed
-
- Bähler J, Wu JQ, Longtine MS, et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. - PubMed
-
- Baijal U, Mehrotra BS. 1965. Species of Mucor from India II. Sydowia 19: 204–212.
-
- Benny GL. 2008. Methods used by Dr. R.K. Benjamin, and other mycologists, to isolate Zygomycetes. Aliso 26: 37–61.
-
- Benny GL, Benjamin RK. 1975. Observations on Thamnidiaceae (Mucorales). New taxa, new combination, and notes on selected species. Aliso 8: 301–351.
LinkOut - more resources
Full Text Sources
Research Materials
