Structure-Guided Engineering of a Complement Component C3-Binding Nanobody Improves Specificity and Adds Cofactor Activity
- PMID: 35935935
- PMCID: PMC9352930
- DOI: 10.3389/fimmu.2022.872536
Structure-Guided Engineering of a Complement Component C3-Binding Nanobody Improves Specificity and Adds Cofactor Activity
Abstract
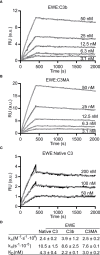
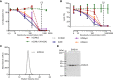
The complement system is a part of the innate immune system, where it labels intruding pathogens as well as dying host cells for clearance. If complement regulation is compromised, the system may contribute to pathogenesis. The proteolytic fragment C3b of complement component C3, is the pivot point of the complement system and provides a scaffold for the assembly of the alternative pathway C3 convertase that greatly amplifies the initial complement activation. This makes C3b an attractive therapeutic target. We previously described a nanobody, hC3Nb1 binding to C3 and its degradation products. Here we show, that extending the N-terminus of hC3Nb1 by a Glu-Trp-Glu motif renders the resulting EWE-hC3Nb1 (EWE) nanobody specific for C3 degradation products. By fusing EWE to N-terminal CCP domains from complement Factor H (FH), we generated the fusion proteins EWEnH and EWEµH. In contrast to EWE, these fusion proteins supported Factor I (FI)-mediated cleavage of human and rat C3b. The EWE, EWEµH, and EWEnH proteins bound C3b and iC3b with low nanomolar dissociation constants and exerted strong inhibition of alternative pathway-mediated deposition of complement. Interestingly, EWEnH remained soluble above 20 mg/mL. Combined with the observed reactivity with both human and rat C3b as well as the ability to support FI-mediated cleavage of C3b, this features EWEnH as a promising candidate for in vivo studies in rodent models of complement driven pathogenesis.
Keywords: Factor H; alternative pathway; complement system; inhibitor; single-domain antibody.
Copyright © 2022 Pedersen, Jensen, Hansen, Petersen, Thiel, Laursen and Andersen.
Conflict of interest statement
Authors HP, RJ, NL, ST and GA are listed as inventors on a patent describing the use of EWE, hC3Nb2 and hC3Nb3. Authors HP, RJ, NL and GA have filed the patent application P6053EP00 for the use of EWEµH and EWEnH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway.J Immunol. 1998 Jun 1;160(11):5596-604. J Immunol. 1998. PMID: 9605165
-
A C3-specific nanobody that blocks all three activation pathways in the human and murine complement system.J Biol Chem. 2020 Jun 26;295(26):8746-8758. doi: 10.1074/jbc.RA119.012339. Epub 2020 May 6. J Biol Chem. 2020. PMID: 32376685 Free PMC article.
-
The Sez6 Family Inhibits Complement by Facilitating Factor I Cleavage of C3b and Accelerating the Decay of C3 Convertases.Front Immunol. 2021 Apr 15;12:607641. doi: 10.3389/fimmu.2021.607641. eCollection 2021. Front Immunol. 2021. PMID: 33936031 Free PMC article.
-
Autoantibodies Against C3b-Functional Consequences and Disease Relevance.Front Immunol. 2019 Jan 29;10:64. doi: 10.3389/fimmu.2019.00064. eCollection 2019. Front Immunol. 2019. PMID: 30761135 Free PMC article. Review.
-
Structure/function of C5 convertases of complement.Int Immunopharmacol. 2001 Mar;1(3):415-22. doi: 10.1016/s1567-5769(00)00039-4. Int Immunopharmacol. 2001. PMID: 11367526 Review.
Cited by
-
Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.Protein Sci. 2022 Oct;31(10):e4432. doi: 10.1002/pro.4432. Protein Sci. 2022. PMID: 36173177 Free PMC article.
-
Modulating the complement system through epitope-specific inhibition by complement C3 inhibitors.J Biol Chem. 2025 Mar;301(3):108250. doi: 10.1016/j.jbc.2025.108250. Epub 2025 Jan 31. J Biol Chem. 2025. PMID: 39894217 Free PMC article.
-
Cryo-EM analysis of complement C3 reveals a reversible major opening of the macroglobulin ring.Nat Struct Mol Biol. 2025 May;32(5):884-895. doi: 10.1038/s41594-024-01467-4. Epub 2025 Jan 23. Nat Struct Mol Biol. 2025. PMID: 39849196
-
Filtration and tubular handling of EWE-hC3Nb1, a complement inhibitor nanobody, in wild type mice and a mouse model of proteinuric kidney disease.FEBS Open Bio. 2024 Feb;14(2):322-330. doi: 10.1002/2211-5463.13752. Epub 2024 Jan 4. FEBS Open Bio. 2024. PMID: 38124617 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

