Epigenetic regulation and T-cell responses in endometriosis - something other than autoimmunity
- PMID: 35935991
- PMCID: PMC9355085
- DOI: 10.3389/fimmu.2022.943839
Epigenetic regulation and T-cell responses in endometriosis - something other than autoimmunity
Abstract
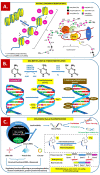
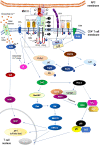
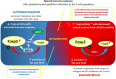
Endometriosis is defined as the presence of endometrial-like glands and stroma located outside the uterine cavity. This common, estrogen dependent, inflammatory condition affects up to 15% of reproductive-aged women and is a well-recognized cause of chronic pelvic pain and infertility. Despite the still unknown etiology of endometriosis, much evidence suggests the participation of epigenetic mechanisms in the disease etiopathogenesis. The main rationale is based on the fact that heritable phenotype changes that do not involve alterations in the DNA sequence are common triggers for hormonal, immunological, and inflammatory disorders, which play a key role in the formation of endometriotic foci. Epigenetic mechanisms regulating T-cell responses, including DNA methylation and posttranslational histone modifications, deserve attention because tissue-resident T lymphocytes work in concert with organ structural cells to generate appropriate immune responses and are functionally shaped by organ-specific environmental conditions. Thus, a failure to precisely regulate immune cell transcription may result in compromised immunological integrity of the organ with an increased risk of inflammatory disorders. The coexistence of endometriosis and autoimmunity is a well-known occurrence. Recent research results indicate regulatory T-cell (Treg) alterations in endometriosis, and an increased number of highly active Tregs and macrophages have been found in peritoneal fluid from women with endometriosis. Elimination of the regulatory function of T cells and an imbalance between T helper cells of the Th1 and Th2 types have been reported in the endometria of women with endometriosis-associated infertility. This review aims to present the state of the art in recognition epigenetic reprogramming of T cells as the key factor in the pathophysiology of endometriosis in the context of T-cell-related autoimmunity. The new potential therapeutic approaches based on epigenetic modulation and/or adoptive transfer of T cells will also be outlined.
Keywords: T cells; T-cell reprogramming; autoimmunity; endometriosis; epigenetic mechanisms.
Copyright © 2022 Szukiewicz.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor APG declared a past co-authorship with the author.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

