IFN-I inducible miR-3614-5p targets ADAR1 isoforms and fine tunes innate immune activation
- PMID: 35935998
- PMCID: PMC9354889
- DOI: 10.3389/fimmu.2022.939907
IFN-I inducible miR-3614-5p targets ADAR1 isoforms and fine tunes innate immune activation
Abstract
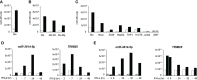
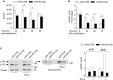
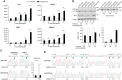
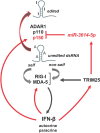
Regulation of innate immune responses is essential for maintenance of immune homeostasis and development of an appropriate immunity against microbial infection. We show here that miR-3614-5p, product of the TRIM25 host gene, is induced by type I interferon (IFN-I) in several human non-immune and immune cell types, in particular in primary myeloid cells. Studies in HeLa cells showed that miR-3614-5p represses both p110 and p150 ADAR1 and reduces constitutive and IFN-induced A-to-I RNA editing. In line with this, activation of innate sensors and expression of IFN-β and the pro-inflammatory IL-6 are promoted. MiR-3614-5p directly targets ADAR1 transcripts by binding to one specific site in the 3'UTR. Moreover, we could show that endogenous miR-3614-5p is associated with Ago2 and targets ADAR1 in IFN-stimulated cells. Overall, we propose that, by reducing ADAR1, IFN-I-induced miR-3614-5p contributes to lowering the activation threshold of innate sensors. Our findings provide new insights into the role of miR-3614-5p, placing it as a potential fine tuner of dsRNA metabolism, cell homeostasis and innate immunity.
Keywords: ADAR1; TRIM25; innate response; microRNA; type I interferon.
Copyright © 2022 Vuillier, Li, Black, Cruciani, Rubino, Michel and Pellegrini.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships.
Figures



Similar articles
-
Ubiquitin-dependent Turnover of Adenosine Deaminase Acting on RNA 1 (ADAR1) Is Required for Efficient Antiviral Activity of Type I Interferon.J Biol Chem. 2016 Nov 25;291(48):24974-24985. doi: 10.1074/jbc.M116.737098. Epub 2016 Oct 11. J Biol Chem. 2016. PMID: 27729454 Free PMC article.
-
Editing of Cellular Self-RNAs by Adenosine Deaminase ADAR1 Suppresses Innate Immune Stress Responses.J Biol Chem. 2016 Mar 18;291(12):6158-68. doi: 10.1074/jbc.M115.709014. Epub 2016 Jan 27. J Biol Chem. 2016. PMID: 26817845 Free PMC article.
-
Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150.PLoS Biol. 2018 Nov 29;16(11):e2006577. doi: 10.1371/journal.pbio.2006577. eCollection 2018 Nov. PLoS Biol. 2018. PMID: 30496178 Free PMC article.
-
An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1).J Interferon Cytokine Res. 2014 Jun;34(6):437-46. doi: 10.1089/jir.2014.0001. J Interferon Cytokine Res. 2014. PMID: 24905200 Free PMC article. Review.
-
Deciphering the Biological Significance of ADAR1-Z-RNA Interactions.Int J Mol Sci. 2021 Oct 23;22(21):11435. doi: 10.3390/ijms222111435. Int J Mol Sci. 2021. PMID: 34768866 Free PMC article. Review.
Cited by
-
A Wonderful Journey: The Diverse Roles of Adenosine Deaminase Action on RNA 1 (ADAR1) in Central Nervous System Diseases.CNS Neurosci Ther. 2025 Jan;31(1):e70208. doi: 10.1111/cns.70208. CNS Neurosci Ther. 2025. PMID: 39753993 Free PMC article. Review.
-
Dual Role of microRNA-146a in Experimental Inflammation in Human Pulmonary Epithelial and Immune Cells and Expression in Inflammatory Lung Diseases.Int J Mol Sci. 2024 Jul 13;25(14):7686. doi: 10.3390/ijms25147686. Int J Mol Sci. 2024. PMID: 39062931 Free PMC article.
-
T cell dysfunction in elderly ARDS patients based on miRNA and mRNA integration analysis.Front Immunol. 2024 Mar 20;15:1368446. doi: 10.3389/fimmu.2024.1368446. eCollection 2024. Front Immunol. 2024. PMID: 38571958 Free PMC article.
-
The Function of TRIM25 in Antiviral Defense and Viral Immune Evasion.Viruses. 2025 May 20;17(5):735. doi: 10.3390/v17050735. Viruses. 2025. PMID: 40431746 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

