Optimizations for Computing Relatedness in Biomedical Heterogeneous Information Networks: SemNet 2.0
- PMID: 35936510
- PMCID: PMC9351549
- DOI: 10.3390/bdcc6010027
Optimizations for Computing Relatedness in Biomedical Heterogeneous Information Networks: SemNet 2.0
Abstract
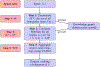
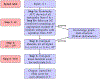
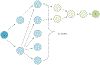
Literature-based discovery (LBD) summarizes information and generates insight from large text corpuses. The SemNet framework utilizes a large heterogeneous information network or "knowledge graph" of nodes and edges to compute relatedness and rank concepts pertinent to a user-specified target. SemNet provides a way to perform multi-factorial and multi-scalar analysis of complex disease etiology and therapeutic identification using the 33+ million articles in PubMed. The present work improves the efficacy and efficiency of LBD for end users by augmenting SemNet to create SemNet 2.0. A custom Python data structure replaced reliance on Neo4j to improve knowledge graph query times by several orders of magnitude. Additionally, two randomized algorithms were built to optimize the HeteSim metric calculation for computing metapath similarity. The unsupervised learning algorithm for rank aggregation (ULARA), which ranks concepts with respect to the user-specified target, was reconstructed using derived mathematical proofs of correctness and probabilistic performance guarantees for optimization. The upgraded ULARA is generalizable to other rank aggregation problems outside of SemNet. In summary, SemNet 2.0 is a comprehensive open-source software for significantly faster, more effective, and user-friendly means of automated biomedical LBD. An example case is performed to rank relationships between Alzheimer's disease and metabolic co-morbidities.
Keywords: Alzheimer’s disease; HeteSim; SemNet; ULARA; biomedical knowledge graph; machine learning; natural language processing; rank aggregation; relatedness; text mining.
Figures

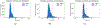
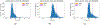
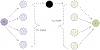
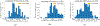
Similar articles
-
CompositeView: A Network-Based Visualization Tool.Big Data Cogn Comput. 2022 Jun;6(2):66. doi: 10.3390/bdcc6020066. Epub 2022 Jun 14. Big Data Cogn Comput. 2022. PMID: 35847767 Free PMC article.
-
SemNet: Using Local Features to Navigate the Biomedical Concept Graph.Front Bioeng Biotechnol. 2019 Jul 3;7:156. doi: 10.3389/fbioe.2019.00156. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31334227 Free PMC article.
-
Serial KinderMiner (SKiM) discovers and annotates biomedical knowledge using co-occurrence and transformer models.BMC Bioinformatics. 2023 Nov 1;24(1):412. doi: 10.1186/s12859-023-05539-y. BMC Bioinformatics. 2023. PMID: 37915001 Free PMC article.
-
Computational Literature-based Discovery for Natural Products Research: Current State and Future Prospects.Front Bioinform. 2022 Mar 15;2:827207. doi: 10.3389/fbinf.2022.827207. eCollection 2022. Front Bioinform. 2022. PMID: 36304281 Free PMC article. Review.
-
Informatics Support for Basic Research in Biomedicine.ILAR J. 2017 Jul 1;58(1):80-89. doi: 10.1093/ilar/ilx004. ILAR J. 2017. PMID: 28838071 Free PMC article. Review.
Cited by
-
Literature-Based Discovery to Elucidate the Biological Links between Resistant Hypertension and COVID-19.Biology (Basel). 2023 Sep 21;12(9):1269. doi: 10.3390/biology12091269. Biology (Basel). 2023. PMID: 37759668 Free PMC article.
-
CompositeView: A Network-Based Visualization Tool.Big Data Cogn Comput. 2022 Jun;6(2):66. doi: 10.3390/bdcc6020066. Epub 2022 Jun 14. Big Data Cogn Comput. 2022. PMID: 35847767 Free PMC article.
-
Artificial Intelligence-Assisted Comparative Analysis of the Overlapping Molecular Pathophysiology of Alzheimer's Disease, Amyotrophic Lateral Sclerosis, and Frontotemporal Dementia.Int J Mol Sci. 2024 Dec 15;25(24):13450. doi: 10.3390/ijms252413450. Int J Mol Sci. 2024. PMID: 39769215 Free PMC article.
-
Cross-Domain Text Mining to Predict Adverse Events from Tyrosine Kinase Inhibitors for Chronic Myeloid Leukemia.Cancers (Basel). 2022 Sep 26;14(19):4686. doi: 10.3390/cancers14194686. Cancers (Basel). 2022. PMID: 36230609 Free PMC article.
-
Natural language processing in Alzheimer's disease research: Systematic review of methods, data, and efficacy.Alzheimers Dement (Amst). 2025 Feb 11;17(1):e70082. doi: 10.1002/dad2.70082. eCollection 2025 Jan-Mar. Alzheimers Dement (Amst). 2025. PMID: 39935888 Free PMC article. Review.
References
-
- PubMed Overview. Available online: https://pubmed.ncbi.nlm.nih.gov/about/ (accessed on 10 November 2021).
-
- Swanson D Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspect. Biol. Med 1986, 30, 7–18. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
