Identification of ankyrin-transmembrane-type subfamily genes in Triticeae species reveals TaANKTM2A-5 regulates powdery mildew resistance in wheat
- PMID: 35937376
- PMCID: PMC9353636
- DOI: 10.3389/fpls.2022.943217
Identification of ankyrin-transmembrane-type subfamily genes in Triticeae species reveals TaANKTM2A-5 regulates powdery mildew resistance in wheat
Abstract
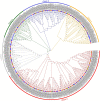
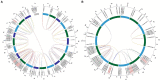
The ankyrin-transmembrane (ANKTM) subfamily is the most abundant subgroup of the ANK superfamily, with critical roles in pathogen defense. However, the function of ANKTM proteins in wheat immunity remains largely unexplored. Here, a total of 381 ANKTMs were identified from five Triticeae species and Arabidopsis, constituting five classes. Among them, class a only contains proteins from Triticeae species and the number of ANKTM in class a of wheat is significantly larger than expected, even after consideration of the ploidy level. Tandem duplication analysis of ANKTM indicates that Triticum urartu, Triticum dicoccoides and wheat all had experienced tandem duplication events which in wheat-produced ANKTM genes all clustered in class a. The above suggests that not only did the genome polyploidization result in the increase of ANKTM gene number, but that tandem duplication is also a mechanism for the expansion of this subfamily. Micro-collinearity analysis of Triticeae ANKTMs indicates that some ANKTM type genes evolved into other types of ANKs in the evolution process. Public RNA-seq data showed that most of the genes in class d and class e are expressed, and some of them show differential responses to biotic stresses. Furthermore, qRT-PCR results showed that some ANKTMs in class d and class e responded to powdery mildew. Silencing of TaANKTM2A-5 by barley stripe mosaic virus-induced gene silencing compromised powdery mildew resistance in common wheat Bainongaikang58. Findings in this study not only help to understand the evolutionary process of ANKTM genes, but also form the basis for exploring disease resistance genes in the ANKTM gene family.
Keywords: ankyrin-transmembrane protein; evolutionary progress; powdery mildew; virus-induced gene silencing; wheat.
Copyright © 2022 Hu, Ren, Xu, Wei, Song, Guan, Gao, Zhang, Hu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Genome-wide identification of TaeGRASs responsive to biotic stresses and functional analysis of TaeSCL6 in wheat resistance to powdery mildew.BMC Genomics. 2024 Nov 27;25(1):1149. doi: 10.1186/s12864-024-11041-3. BMC Genomics. 2024. PMID: 39604842 Free PMC article.
-
Identification of acyl-CoA-binding protein gene in Triticeae species reveals that TaACBP4A-1 and TaACBP4A-2 positively regulate powdery mildew resistance in wheat.Int J Biol Macromol. 2023 Aug 15;246:125526. doi: 10.1016/j.ijbiomac.2023.125526. Epub 2023 Jun 26. Int J Biol Macromol. 2023. PMID: 37379955
-
Genome-Wide Analysis of Serine Hydroxymethyltransferase Genes in Triticeae Species Reveals That TaSHMT3A-1 Regulates Fusarium Head Blight Resistance in Wheat.Front Plant Sci. 2022 Feb 10;13:847087. doi: 10.3389/fpls.2022.847087. eCollection 2022. Front Plant Sci. 2022. PMID: 35222497 Free PMC article.
-
A newly identified photosystem II Subunit P gene TaPsbP4A-1 in Triticeae species negatively regulates wheat powdery mildew resistance.Front Plant Sci. 2024 Nov 8;15:1452281. doi: 10.3389/fpls.2024.1452281. eCollection 2024. Front Plant Sci. 2024. PMID: 39582632 Free PMC article.
-
Genome-wide identification and characterization of the Lateral Organ Boundaries Domain (LBD) gene family in polyploid wheat and related species.PeerJ. 2021 Aug 11;9:e11811. doi: 10.7717/peerj.11811. eCollection 2021. PeerJ. 2021. PMID: 34447619 Free PMC article.
Cited by
-
Genome-wide identification of HSP90 gene family in Rosa chinensis and its response to salt and drought stresses.3 Biotech. 2024 Sep;14(9):204. doi: 10.1007/s13205-024-04052-0. Epub 2024 Aug 18. 3 Biotech. 2024. PMID: 39161880 Free PMC article.
-
Cellular Partners of Tobamoviral Movement Proteins.Int J Mol Sci. 2025 Jan 5;26(1):400. doi: 10.3390/ijms26010400. Int J Mol Sci. 2025. PMID: 39796254 Free PMC article. Review.
-
GmANKTM21 Positively Regulates Drought Tolerance and Enhanced Stomatal Response through the MAPK Signaling Pathway in Soybean.Int J Mol Sci. 2024 Jun 26;25(13):6972. doi: 10.3390/ijms25136972. Int J Mol Sci. 2024. PMID: 39000082 Free PMC article.
-
Genome-Wide Association Study of Seed Morphology Traits in Senegalese Sorghum Cultivars.Plants (Basel). 2023 Jun 16;12(12):2344. doi: 10.3390/plants12122344. Plants (Basel). 2023. PMID: 37375969 Free PMC article.
References
-
- Chen Y., Song W., Xie X., Wang Z., Guan P., Peng H., et al. . (2020b). A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the Triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant 13, 1694–1708. doi: 10.1016/j.molp.2020.09.019, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

