Correlations between Intestinal Microbiota and Clinical Characteristics in Colorectal Adenoma/Carcinoma
- PMID: 35937408
- PMCID: PMC9352470
- DOI: 10.1155/2022/3140070
Correlations between Intestinal Microbiota and Clinical Characteristics in Colorectal Adenoma/Carcinoma
Retraction in
-
Retracted: Correlations between Intestinal Microbiota and Clinical Characteristics in Colorectal Adenoma/Carcinoma.Biomed Res Int. 2023 Dec 29;2023:9810254. doi: 10.1155/2023/9810254. eCollection 2023. Biomed Res Int. 2023. PMID: 38188806 Free PMC article.
Abstract
Background: Most of colorectal cancer (CRC) cases are sporadic and develop along the adenoma-carcinoma sequence. Intestinal microbial dysbiosis is involved in the development of colorectal cancer. However, there are still no absolute markers predicting the progression from adenoma to carcinoma.
Aims: To investigate the characteristics of intestinal microbiota in colorectal adenoma and carcinoma patients and the correlations with clinical characteristics.
Methods: Fecal samples were collected from 154 colorectal carcinoma patients (CRC group), 20 colorectal adenoma patients (AD group), and 199 healthy controls (control group). The intestinal microbiota was investigated by 16S rRNA gene sequencing.
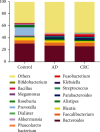
Results: Compared to the healthy controls, microbial diversity was dramatically decreased in AD/CRC. At the genus level, Acidaminococcus significantly decreased with the order of control-AD-CRC (P < 0.05). Parvimonas, Peptostreptococcus, Prevotella, Butyricimonas, Alistipes, and Odoribacter were the key genera in the network of colorectal adenoma/carcinoma-associated bacteria. Combination of the top 10 most important species, including Butyricimonas synergistica, Agrobacterium larrymoorei, Bacteroides plebeius, Lachnospiraceae bacterium feline oral taxon 001, Clostridium scindens, Prevotella heparinolytica, bacterium LD2013, Streptococcus mutans, Lachnospiraceae bacterium 19gly4, and Eubacterium hallii, showed the best performance in distinguishing AD patients from CRC (AUC = 85.54%, 95% CI: 78.83%-92.25%). The clinicopathologic features, including age, sex, tumor location, differentiation degree, and TNM stage, were identified to be closely linked to the intestinal microbiome in CRC.
Conclusion: Several intestinal bacteria changed along the adenoma-carcinoma sequence and might be the potential markers for the diagnosis and treatment of colorectal adenoma/carcinoma. Intestinal microbiota characteristics in CRC should account for the host factors.
Copyright © 2022 Caizhao Lin et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

