Identification of the ubiquitin-proteasome pathway domain by hyperparameter optimization based on a 2D convolutional neural network
- PMID: 35937990
- PMCID: PMC9355632
- DOI: 10.3389/fgene.2022.851688
Identification of the ubiquitin-proteasome pathway domain by hyperparameter optimization based on a 2D convolutional neural network
Abstract
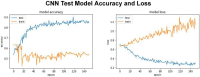
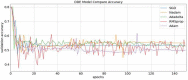
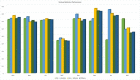
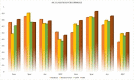
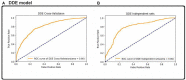
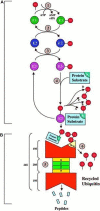
The major mechanism of proteolysis in the cytosol and nucleus is the ubiquitin-proteasome pathway (UPP). The highly controlled UPP has an effect on a wide range of cellular processes and substrates, and flaws in the system can lead to the pathogenesis of a number of serious human diseases. Knowledge about UPPs provide useful hints to understand the cellular process and drug discovery. The exponential growth in next-generation sequencing wet lab approaches have accelerated the accumulation of unannotated data in online databases, making the UPP characterization/analysis task more challenging. Thus, computational methods are used as an alternative for fast and accurate identification of UPPs. Aiming this, we develop a novel deep learning-based predictor named "2DCNN-UPP" for identifying UPPs with low error rate. In the proposed method, we used proposed algorithm with a two-dimensional convolutional neural network with dipeptide deviation features. To avoid the over fitting problem, genetic algorithm is employed to select the optimal features. Finally, the optimized attribute set are fed as input to the 2D-CNN learning engine for building the model. Empirical evidence or outcomes demonstrates that the proposed predictor achieved an overall accuracy and AUC (ROC) value using 10-fold cross validation test. Superior performance compared to other state-of-the art methods for discrimination the relations UPPs classification. Both on and independent test respectively was trained on 10-fold cross validation method and then evaluated through independent test. In the case where experimentally validated ubiquitination sites emerged, we must devise a proteomics-based predictor of ubiquitination. Meanwhile, we also evaluated the generalization power of our trained modal via independent test, and obtained remarkable performance in term of 0.862 accuracy, 0.921 sensitivity, 0.803 specificity 0.803, and 0.730 Matthews correlation coefficient (MCC) respectively. Four approaches were used in the sequences, and the physical properties were calculated combined. When used a 10-fold cross-validation, 2D-CNN-UPP obtained an AUC (ROC) value of 0.862 predicted score. We analyzed the relationship between UPP protein and non-UPP protein predicted score. Last but not least, this research could effectively analyze the large scale relationship between UPP proteins and non-UPP proteins in particular and other protein problems in general and our research work might improve computational biological research. Therefore, we could utilize the latest features in our model framework and Dipeptide Deviation from Expected Mean (DDE) -based protein structure features for the prediction of protein structure, functions, and different molecules, such as DNA and RNA.
Keywords: 2D-CNN; CNN; DDE; protein sequence prediction; ubiquitin-proteasome pathway.
Copyright © 2022 Sikander, Arif, Ghulam, Worachartcheewan, Thafar and Habib.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


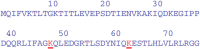


References
-
- Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., et al. (2015). TensorFlow: Large-Scale machine learning on heterogeneous systems. Available from: https://www.tensorflow.org/ .
-
- Abdel-Hamid O., Deng L., Yu D. (2013). Exploring convolutional neural network structures and optimization techniques for speech recognition. Interspeech 11, 73–75. 10.21437/interspeech.2013-744 - DOI
-
- Bergstra J., Bardenet R., Bengio Y., Kégl B. (2011). “Algorithms for hyper-parameter optimization,” in Advances in neural information processing systems, 24.
-
- Billones C. D., Demetria O. J. L. D., Hostallero D. E. D., Naval P. C. (2016). “DemNet: A convolutional neural network for the detection of alzheimer's disease and mild cognitive impairment,” in Proceedings of the 2016 IEEE region 10 conference (TENCON), Singapore, November 2016 (IEEE; ), 3724–3727. 10.1109/tencon.2016.7848755 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

