Profiling a Community-Specific Function Landscape for Bacterial Peptides Through Protein-Level Meta-Assembly and Machine Learning
- PMID: 35938008
- PMCID: PMC9354662
- DOI: 10.3389/fgene.2022.935351
Profiling a Community-Specific Function Landscape for Bacterial Peptides Through Protein-Level Meta-Assembly and Machine Learning
Abstract
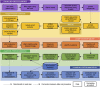
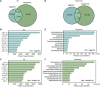
Small proteins, encoded by small open reading frames, are only beginning to emerge with the current advancement of omics technology and bioinformatics. There is increasing evidence that small proteins play roles in diverse critical biological functions, such as adjusting cellular metabolism, regulating other protein activities, controlling cell cycles, and affecting disease physiology. In prokaryotes such as bacteria, the small proteins are largely unexplored for their sequence space and functional groups. For most bacterial species from a natural community, the sample cannot be easily isolated or cultured, and the bacterial peptides must be better characterized in a metagenomic manner. The bacterial peptides identified from metagenomic samples can not only enrich the pool of small proteins but can also reveal the community-specific microbe ecology information from a small protein perspective. In this study, metaBP (Bacterial Peptides for metagenomic sample) has been developed as a comprehensive toolkit to explore the small protein universe from metagenomic samples. It takes raw sequencing reads as input, performs protein-level meta-assembly, and computes bacterial peptide homolog groups with sample-specific mutations. The metaBP also integrates general protein annotation tools as well as our small protein-specific machine learning module metaBP-ML to construct a full landscape for bacterial peptides. The metaBP-ML shows advantages for discovering functions of bacterial peptides in a microbial community and increases the yields of annotations by up to five folds. The metaBP toolkit demonstrates its novelty in adopting the protein-level assembly to discover small proteins, integrating protein-clustering tool in a new and flexible environment of RBiotools, and presenting the first-time small protein landscape by metaBP-ML. Taken together, metaBP (and metaBP-ML) can profile functional bacterial peptides from metagenomic samples with potential diverse mutations, in order to depict a unique landscape of small proteins from a microbial community.
Keywords: bacterial peptide; machine learning; metagenomics; protein annotation; protein clustering.
Copyright © 2022 Vajjala, Johnson, Kasparek, Leuze and Yao.
Conflict of interest statement
ML is affiliated with Nashville Biosciences. His development of the RBiotools package was done independently of any funding from Nashville Biosciences or from any other commercial funding source. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A multi-source domain annotation pipeline for quantitative metagenomic and metatranscriptomic functional profiling.Microbiome. 2018 Aug 28;6(1):149. doi: 10.1186/s40168-018-0532-2. Microbiome. 2018. PMID: 30153857 Free PMC article.
-
The future of Cochrane Neonatal.Early Hum Dev. 2020 Nov;150:105191. doi: 10.1016/j.earlhumdev.2020.105191. Epub 2020 Sep 12. Early Hum Dev. 2020. PMID: 33036834
-
Massive metagenomic data analysis using abundance-based machine learning.Biol Direct. 2019 Aug 1;14(1):12. doi: 10.1186/s13062-019-0242-0. Biol Direct. 2019. PMID: 31370905 Free PMC article.
-
Machine Learning and Deep Learning Applications in Metagenomic Taxonomy and Functional Annotation.Front Microbiol. 2022 Mar 14;13:811495. doi: 10.3389/fmicb.2022.811495. eCollection 2022. Front Microbiol. 2022. PMID: 35359727 Free PMC article. Review.
-
Metagenomics and Bioinformatics in Microbial Ecology: Current Status and Beyond.Microbes Environ. 2016 Sep 29;31(3):204-12. doi: 10.1264/jsme2.ME16024. Epub 2016 Jul 5. Microbes Environ. 2016. PMID: 27383682 Free PMC article. Review.
Cited by
-
SProtFP: a machine learning-based method for functional classification of small ORFs in prokaryotes.NAR Genom Bioinform. 2025 Jan 7;7(1):lqae186. doi: 10.1093/nargab/lqae186. eCollection 2025 Mar. NAR Genom Bioinform. 2025. PMID: 39781515 Free PMC article.
-
A survey of experimental and computational identification of small proteins.Brief Bioinform. 2024 May 23;25(4):bbae345. doi: 10.1093/bib/bbae345. Brief Bioinform. 2024. PMID: 39007598 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

