Single-cell transcriptomics reveals male germ cells and Sertoli cells developmental patterns in dairy goats
- PMID: 35938151
- PMCID: PMC9355508
- DOI: 10.3389/fcell.2022.944325
Single-cell transcriptomics reveals male germ cells and Sertoli cells developmental patterns in dairy goats
Abstract
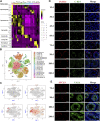
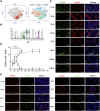
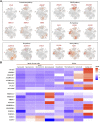
Spermatogenesis holds considerable promise for human-assisted reproduction and livestock breeding based on stem cells. It occurs in seminiferous tubules within the testis, which mainly comprise male germ cells and Sertoli cells. While the developmental progression of male germ cells and Sertoli cells has been widely reported in mice, much less is known in other large animal species, including dairy goats. In this study, we present the data of single cell RNA sequencing (scRNA-seq) for 25,373 cells from 45 (pre-puberty), 90 (puberty), and 180-day-old (post-puberty) dairy goat testes. We aimed to identify genes that are associated with key developmental events in male germ cells and Sertoli cells. We examined the development of spermatogenic cells and seminiferous tubules from 15, 30, 45, 60, 75, 90, 180, and 240-day-old buck goat testes. scRNA-seq clustering analysis of testicular cells from pre-puberty, puberty, and post-puberty goat testes revealed several cell types, including cell populations with characteristics of spermatogonia, early spermatocytes, spermatocytes, spermatids, Sertoli cells, Leydig cells, macrophages, and endothelial cells. We mapped the timeline for male germ cells development from spermatogonia to spermatids and identified gene signatures that define spermatogenic cell populations, such as AMH, SOHLH1, INHA, and ACTA2. Importantly, using immunofluorescence staining for different marker proteins (UCHL1, C-KIT, VASA, SOX9, AMH, and PCNA), we explored the proliferative activity and development of male germ cells and Sertoli cells. Moreover, we identified the expression patterns of potential key genes associated with the niche-related key pathways in male germ cells of dairy goats, including testosterone, retinoic acid, PDGF, FGF, and WNT pathways. In summary, our study systematically investigated the elaborate male germ cells and Sertoli cells developmental patterns in dairy goats that have so far remained largely unknown. This information represents a valuable resource for the establishment of goat male reproductive stem cells lines, induction of germ cell differentiation in vitro, and the exploration of sequential cell fate transition for spermatogenesis and testicular development at single-cell resolution.
Keywords: Sertoli cell; dairy goat; single-cell RNA sequencing (scRNA-seq); spermatogenesis; testes.
Copyright © 2022 Ren, Xi, Qiao, Li, Xian, Zhu and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

