Novel interaction interfaces mediate the interaction between the NEIL1 DNA glycosylase and mitochondrial transcription factor A
- PMID: 35938152
- PMCID: PMC9354671
- DOI: 10.3389/fcell.2022.893806
Novel interaction interfaces mediate the interaction between the NEIL1 DNA glycosylase and mitochondrial transcription factor A
Abstract
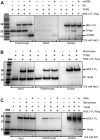
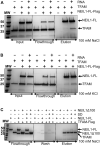
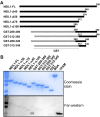
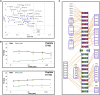
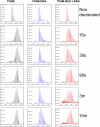
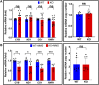
The maintenance of human mitochondrial DNA (mtDNA) is critical for proper cellular function as damage to mtDNA, if left unrepaired, can lead to a diverse array of pathologies. Of the pathways identified to participate in DNA repair within the mitochondria, base excision repair (BER) is the most extensively studied. Protein-protein interactions drive the step-by-step coordination required for the successful completion of this pathway and are important for crosstalk with other mitochondrial factors involved in genome maintenance. Human NEIL1 is one of seven DNA glycosylases that initiates BER in both the nuclear and mitochondrial compartments. In the current work, we scrutinized the interaction between NEIL1 and mitochondrial transcription factor A (TFAM), a protein that is essential for various aspects of mtDNA metabolism. We note, for the first time, that both the N- and C- terminal domains of NEIL1 interact with TFAM revealing a unique NEIL1 protein-binding interface. The interaction between the two proteins, as observed biochemically, appears to be transient and is most apparent at concentrations of low salt. The presence of DNA (or RNA) also positively influences the interaction between the two proteins, and molar mass estimates indicate that duplex DNA is required for complex formation at higher salt concentrations. Hydrogen deuterium exchange mass spectrometry data reveal that both proteins exchange less deuterium upon DNA binding, indicative of an interaction, and the addition of NEIL1 to the TFAM-DNA complex alters the interaction landscape. The transcriptional activity of TFAM appears to be independent of NEIL1 expression under normal cellular conditions, however, in the presence of DNA damage, we observe a significant reduction in the mRNA expression of TFAM-transcribed mitochondrial genes in the absence of NEIL1. Overall, our data indicate that the interaction between NEIL1 and TFAM can be modulated by local environment such as salt concentrations, protein availability, the presence of nucleic acids, as well as the presence of DNA damage.
Keywords: HDX-MS (hydrogen–deuterium exchange mass-spectrometry); NEIL1 DNA glycosylase; base excision repair (BER); mitochondrial transcription factor A (TFAM); small angle X-ray scattering (SAXS).
Copyright © 2022 Sharma, Thompson, Arrington, Terry, Chakravarthy, Prevelige and Prakash.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
The C-terminal tail of the NEIL1 DNA glycosylase interacts with the human mitochondrial single-stranded DNA binding protein.DNA Repair (Amst). 2018 May;65:11-19. doi: 10.1016/j.dnarep.2018.02.012. Epub 2018 Mar 6. DNA Repair (Amst). 2018. PMID: 29522991 Free PMC article.
-
The C-terminal Domain (CTD) of Human DNA Glycosylase NEIL1 Is Required for Forming BERosome Repair Complex with DNA Replication Proteins at the Replicating Genome: DOMINANT NEGATIVE FUNCTION OF THE CTD.J Biol Chem. 2015 Aug 21;290(34):20919-20933. doi: 10.1074/jbc.M115.642918. Epub 2015 Jul 1. J Biol Chem. 2015. PMID: 26134572 Free PMC article.
-
Destabilization of the PCNA trimer mediated by its interaction with the NEIL1 DNA glycosylase.Nucleic Acids Res. 2017 Mar 17;45(5):2897-2909. doi: 10.1093/nar/gkw1282. Nucleic Acids Res. 2017. PMID: 27994037 Free PMC article.
-
Decoding the Role of NEIL1 Gene in DNA Repair and Lifespan: A Literature Review with Bioinformatics Analysis.Adv Biol (Weinh). 2024 Nov;8(11):e2300708. doi: 10.1002/adbi.202300708. Epub 2024 Aug 20. Adv Biol (Weinh). 2024. PMID: 39164210 Review.
-
New paradigms in the repair of oxidative damage in human genome: mechanisms ensuring repair of mutagenic base lesions during replication and involvement of accessory proteins.Cell Mol Life Sci. 2015 May;72(9):1679-98. doi: 10.1007/s00018-014-1820-z. Epub 2015 Jan 10. Cell Mol Life Sci. 2015. PMID: 25575562 Free PMC article. Review.
Cited by
-
NEIL1 block IFN-β production and enhance vRNP function to facilitate influenza A virus proliferation.Npj Viruses. 2024 Nov 21;2(1):57. doi: 10.1038/s44298-024-00065-x. Npj Viruses. 2024. PMID: 40295715 Free PMC article.
-
Examining DNA Structures with In-droplet Hydrogen/Deuterium Exchange Mass Spectrometry.Int J Mass Spectrom. 2024 May;499:117231. doi: 10.1016/j.ijms.2024.117231. Epub 2024 Mar 7. Int J Mass Spectrom. 2024. PMID: 38854816 Free PMC article.
-
Deciphering the crystal structure of a novel nanobody against the NEIL1 DNA glycosylase.Acta Crystallogr D Struct Biol. 2024 Feb 1;80(Pt 2):137-146. doi: 10.1107/S205979832400038X. Epub 2024 Jan 30. Acta Crystallogr D Struct Biol. 2024. PMID: 38289715 Free PMC article.
-
Frequencies and spectra of aflatoxin B1-induced mutations in liver genomes of NEIL1-deficient mice as revealed by duplex sequencing.NAR Mol Med. 2024 May 17;1(2):ugae006. doi: 10.1093/narmme/ugae006. eCollection 2024 Apr. NAR Mol Med. 2024. PMID: 38779538 Free PMC article.
-
Development and characterization of a novel NEIL1 nanobody.DNA Repair (Amst). 2025 Jun;150:103849. doi: 10.1016/j.dnarep.2025.103849. Epub 2025 May 22. DNA Repair (Amst). 2025. PMID: 40440929
References
Grants and funding
LinkOut - more resources
Full Text Sources

