New insights into the evolutionary dynamic and lineage divergence of gasdermin E in metazoa
- PMID: 35938154
- PMCID: PMC9355259
- DOI: 10.3389/fcell.2022.952015
New insights into the evolutionary dynamic and lineage divergence of gasdermin E in metazoa
Abstract
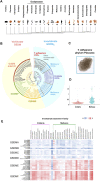
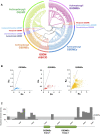
Gasdermin (GSDM) is a family of pore-forming proteins that induce pyroptosis. To date, the origin and evolution of GSDM in Metazoa remain elusive. Here, we found that GSDM emerged early in Placozoa but is absent in a large number of invertebrates. In the lower vertebrate, fish, three types of GSDME, i.e., GSDMEa, GSDMEb, and a previously unreported type (designated GSDMEc), were idenitied. Evolutionarily, the three GSDMEs are distinctly separated: GSDMEa is closely related to tetrapod GSDME; GSDMEb exists exclusively in fish; GSDMEc forms the lineage root of tetrapod GSDMA/B/C/D. GSDMEc shares conserved genomic features with and is probably the prototype of GSDMA, which we found existing in all tetrapod classes. GSDMEc displays fast evolutionary dynamics, likely as a result of genomic transposition. A cross-metazoan analysis of GSDME revealed that GSDMEa shares a conserved caspase recognition motif with the GSDME of tetrapods and cnidarians, whereas GSDMEb has a unique caspase recognition motif similar to that of mammalian GSDMD, and GSDMEc exhibits no apparent caspase recognition motif. Through functional test, four highly conserved residues in vertebrate GSDME proved to be essential to auto-inhibition. Together our results provide new insights into the origin, evolution, and function of metazoan GSDMs.
Keywords: auto-inhibition; evolution; fish; gasdermin; metazoan; recognition motif.
Copyright © 2022 Yuan, Jiang, Qin and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Angosto-Bazarra D., Alarcon-Vila C., Hurtado-Navarro L., Banos M. C., Rivers-Auty J., Pelegrin P., et al. (2022). Evolutionary analyses of the gasdermin family suggest conserved roles in infection response despite loss of pore-forming functionality. BMC Biol. 20 (1), 9. 10.1186/s12915-021-01220-z - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

