Comprehensive Analysis of Quantitative Proteomics With DIA Mass Spectrometry and ceRNA Network in Intrahepatic Cholestasis of Pregnancy
- PMID: 35938169
- PMCID: PMC9354660
- DOI: 10.3389/fcell.2022.854425
Comprehensive Analysis of Quantitative Proteomics With DIA Mass Spectrometry and ceRNA Network in Intrahepatic Cholestasis of Pregnancy
Abstract
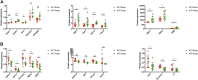
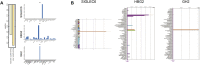
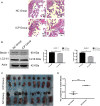
Background: Intrahepatic cholestasis of pregnancy (ICP) is a pregnancy-specific complication characterized by pruritus without skin damage and jaundice. The poor perinatal outcomes include fetal distress, preterm birth, and unexpected intrauterine death. However, the mechanism of ICP leading to poor prognosis is still unclear. Methods: We analyzed 10 ICP and 10 normal placental specimens through quantitative proteomics of data-independent acquisition (DIA) to screen and identify differentially expressed proteins. GO, KEGG, COG/KOG, StringDB, InterProScan, Metascape, BioGPS, and NetworkAnalyst databases were used in this study. PITA, miRanda, TargetScan, starBase, and LncBase Predicted v.2 were used for constructing a competing endogenous RNA (ceRNA) network. Cytoscape was used for drawing regulatory networks, and cytoHubba was used for screening core nodes. The ICP rat models were used to validate the pathological mechanism. Results: GO, KEGG, and COG/KOG functional enrichment analysis results showed the differentially expressed proteins participated in autophagy, autophagosome formation, cofactor binding, JAK-STAT signaling pathway, and coenzyme transport and metabolism. DisGeNET analysis showed that these differentially expressed proteins were associated with red blood cell disorder and slow progression. We further analyzed first 12 proteins in the upregulated and downregulated differentially expressed proteins and incorporated clinicopathologic parameters. Our results showed HBG1, SPI1, HBG2, HBE1, FOXK1, KRT72, SLC13A3, MBD2, SP9, GPLD1, MYH7, and BLOC1S1 were associated with ICP development. ceRNA network analysis showed that MBD2, SPI1, FOXK1, and SLC13A3 were regulated by multiple miRNAs and lncRNAs. Conclusion: ICP was associated with autophagy. The ceRNA network of MBD2, SPI1, FOXK1, and SLC13A3 was involved in ICP progression, and these core proteins might be potential target.
Keywords: competing endogenous RNA (ceRNA) network; intrahepatic cholestasis of pregnancy; quantitative proteomics; regulatory mechanism; target therapy.
Copyright © 2022 Fang, Fang, Zhang, Xiang, Cheng, Liang and Xia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Core biomarkers analysis benefit for diagnosis on human intrahepatic cholestasis of pregnancy.BMC Pregnancy Childbirth. 2024 Aug 10;24(1):525. doi: 10.1186/s12884-024-06730-6. BMC Pregnancy Childbirth. 2024. PMID: 39127651 Free PMC article.
-
Immune Dysfunction Mediated by the ceRNA Regulatory Network in Human Placenta Tissue of Intrahepatic Cholestasis Pregnancy.Front Immunol. 2022 Jun 24;13:883971. doi: 10.3389/fimmu.2022.883971. eCollection 2022. Front Immunol. 2022. PMID: 35812382 Free PMC article.
-
Comprehensive analysis of placental gene-expression profiles and identification of EGFR-mediated autophagy and ferroptosis suppression in intrahepatic cholestasis of pregnancy.Gene. 2022 Aug 5;834:146594. doi: 10.1016/j.gene.2022.146594. Epub 2022 May 25. Gene. 2022. PMID: 35643225
-
Analysis of long noncoding RNA-associated competing endogenous RNA network in glucagon-like peptide-1 receptor agonist-mediated protection in β cells.World J Diabetes. 2020 Sep 15;11(9):374-390. doi: 10.4239/wjd.v11.i9.374. World J Diabetes. 2020. PMID: 32994866 Free PMC article.
-
Evaluating the effectiveness and safety of ursodeoxycholic acid in treatment of intrahepatic cholestasis of pregnancy: A meta-analysis (a prisma-compliant study).Medicine (Baltimore). 2016 Oct;95(40):e4949. doi: 10.1097/MD.0000000000004949. Medicine (Baltimore). 2016. PMID: 27749550 Free PMC article. Review.
Cited by
-
Maternal PM2.5 exposure is associated with preterm birth and gestational diabetes mellitus, and mitochondrial OXPHOS dysfunction in cord blood.Environ Sci Pollut Res Int. 2024 Feb;31(7):10565-10578. doi: 10.1007/s11356-023-31774-0. Epub 2024 Jan 10. Environ Sci Pollut Res Int. 2024. PMID: 38200189 Free PMC article.
-
Core biomarkers analysis benefit for diagnosis on human intrahepatic cholestasis of pregnancy.BMC Pregnancy Childbirth. 2024 Aug 10;24(1):525. doi: 10.1186/s12884-024-06730-6. BMC Pregnancy Childbirth. 2024. PMID: 39127651 Free PMC article.
-
LC-MS/MS untargeted lipidomics uncovers placenta lipid signatures from intrahepatic cholestasis of pregnancy.Front Physiol. 2024 Jun 3;15:1276722. doi: 10.3389/fphys.2024.1276722. eCollection 2024. Front Physiol. 2024. PMID: 38887316 Free PMC article.
-
Integrative proteomics and metabolomics data analysis exploring the mechanism of brain injury after cardiac surgery in chronic stress rats.BMC Anesthesiol. 2024 Mar 22;24(1):111. doi: 10.1186/s12871-024-02492-y. BMC Anesthesiol. 2024. PMID: 38519946 Free PMC article.
-
The role of noncoding RNA and its diagnostic potential in intrahepatic cholestasis of pregnancy: a research update.Front Genet. 2023 Oct 13;14:1239693. doi: 10.3389/fgene.2023.1239693. eCollection 2023. Front Genet. 2023. PMID: 37900174 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

