Transcription Factor GATA4 Regulates Cell Type-Specific Splicing Through Direct Interaction With RNA in Human Induced Pluripotent Stem Cell-Derived Cardiac Progenitors
- PMID: 35938400
- PMCID: PMC9452483
- DOI: 10.1161/CIRCULATIONAHA.121.057620
Transcription Factor GATA4 Regulates Cell Type-Specific Splicing Through Direct Interaction With RNA in Human Induced Pluripotent Stem Cell-Derived Cardiac Progenitors
Abstract
Background: GATA4 (GATA-binding protein 4), a zinc finger-containing, DNA-binding transcription factor, is essential for normal cardiac development and homeostasis in mice and humans, and mutations in this gene have been reported in human heart defects. Defects in alternative splicing are associated with many heart diseases, yet relatively little is known about how cell type- or cell state-specific alternative splicing is achieved in the heart. Here, we show that GATA4 regulates cell type-specific splicing through direct interaction with RNA and the spliceosome in human induced pluripotent stem cell-derived cardiac progenitors.
Methods: We leveraged a combination of unbiased approaches including affinity purification of GATA4 and mass spectrometry, enhanced cross-linking with immunoprecipitation, electrophoretic mobility shift assays, in vitro splicing assays, and unbiased transcriptomic analysis to uncover GATA4's novel function as a splicing regulator in human induced pluripotent stem cell-derived cardiac progenitors.
Results: We found that GATA4 interacts with many members of the spliceosome complex in human induced pluripotent stem cell-derived cardiac progenitors. Enhanced cross-linking with immunoprecipitation demonstrated that GATA4 also directly binds to a large number of mRNAs through defined RNA motifs in a sequence-specific manner. In vitro splicing assays indicated that GATA4 regulates alternative splicing through direct RNA binding, resulting in functionally distinct protein products. Correspondingly, knockdown of GATA4 in human induced pluripotent stem cell-derived cardiac progenitors resulted in differential alternative splicing of genes involved in cytoskeleton organization and calcium ion import, with functional consequences associated with the protein isoforms.
Conclusions: This study shows that in addition to its well described transcriptional function, GATA4 interacts with members of the spliceosome complex and regulates cell type-specific alternative splicing via sequence-specific interactions with RNA. Several genes that have splicing regulated by GATA4 have functional consequences and many are associated with dilated cardiomyopathy, suggesting a novel role for GATA4 in achieving the necessary cardiac proteome in normal and stress-responsive conditions.
Keywords: GATA4 transcription factor; RNA splicing; RNA-binding motifs; induced pluripotent stem cells; myocytes, cardiac.
Conflict of interest statement
Declaration of Interests:
D.S. is a co-founder and member of the board of directors of Tenaya Therapeutics and has equity in Tenaya Therapeutics; B.G.B. and B.R.C. are co-founders with equity in Tenaya Therapeutics; K.S.P. and N.K. have equity in Tenaya Therapeutics. The N.K. Laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. N.K. has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics and Interline Therapeutics and has received stocks from Maze Therapeutics and Interline Therapeutics. The other authors declare no competing interests.
Figures


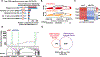

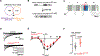

Similar articles
-
RBPMS regulates cardiomyocyte contraction and cardiac function through RNA alternative splicing.Cardiovasc Res. 2024 Feb 27;120(1):56-68. doi: 10.1093/cvr/cvad166. Cardiovasc Res. 2024. PMID: 37890031 Free PMC article.
-
Patient-specific iPSC-derived cardiomyocytes reveal abnormal regulation of FGF16 in a familial atrial septal defect.Cardiovasc Res. 2022 Feb 21;118(3):859-871. doi: 10.1093/cvr/cvab154. Cardiovasc Res. 2022. PMID: 33956078
-
Bicuspid Aortic Valve-Associated Regulatory Regions Reveal GATA4 Regulation and Function During Human-Induced Pluripotent Stem Cell-Based Endothelial-Mesenchymal Transition-Brief Report.Arterioscler Thromb Vasc Biol. 2023 Feb;43(2):312-322. doi: 10.1161/ATVBAHA.122.318566. Epub 2022 Dec 15. Arterioscler Thromb Vasc Biol. 2023. PMID: 36519469 Free PMC article.
-
Towards Understanding the Gene-Specific Roles of GATA Factors in Heart Development: Does GATA4 Lead the Way?Int J Mol Sci. 2022 May 9;23(9):5255. doi: 10.3390/ijms23095255. Int J Mol Sci. 2022. PMID: 35563646 Free PMC article. Review.
-
[Development of Targeted Pharmacotherapy for Cardiovascular Disease].Yakugaku Zasshi. 2017;137(11):1349-1353. doi: 10.1248/yakushi.17-00171. Yakugaku Zasshi. 2017. PMID: 29093370 Review. Japanese.
Cited by
-
Transcription factors and splice factors - interconnected regulators of stem cell differentiation.Curr Stem Cell Rep. 2023 Jun;9(2):31-41. doi: 10.1007/s40778-023-00227-2. Epub 2023 Jun 29. Curr Stem Cell Rep. 2023. PMID: 38939410 Free PMC article.
-
Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.Mol Cell. 2024 Aug 22;84(16):3061-3079.e10. doi: 10.1016/j.molcel.2024.07.016. Epub 2024 Aug 8. Mol Cell. 2024. PMID: 39121853 Free PMC article.
-
RNA binding proteins in cardiovascular development and disease.Curr Top Dev Biol. 2024;156:51-119. doi: 10.1016/bs.ctdb.2024.01.007. Epub 2024 Mar 15. Curr Top Dev Biol. 2024. PMID: 38556427 Free PMC article.
-
Uncovering the Genetic Basis of Congenital Heart Disease: Recent Advancements and Implications for Clinical Management.CJC Pediatr Congenit Heart Dis. 2023 Oct 19;2(6Part B):464-480. doi: 10.1016/j.cjcpc.2023.10.008. eCollection 2023 Dec. CJC Pediatr Congenit Heart Dis. 2023. PMID: 38205435 Free PMC article. Review.
-
The role of GATA4 in mesenchymal stem cell senescence: A new frontier in regenerative medicine.Regen Ther. 2024 Dec 25;28:214-226. doi: 10.1016/j.reth.2024.11.017. eCollection 2025 Mar. Regen Ther. 2024. PMID: 39811069 Free PMC article. Review.
References
-
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science (New York, NY. 2015;350:1262–1266. doi: 10.1126/science.aac9396 - DOI - PMC - PubMed

