The Plasmodium falciparum Nuclear Protein Phosphatase NIF4 Is Required for Efficient Merozoite Invasion and Regulates Artemisinin Sensitivity
- PMID: 35938722
- PMCID: PMC9426563
- DOI: 10.1128/mbio.01897-22
The Plasmodium falciparum Nuclear Protein Phosphatase NIF4 Is Required for Efficient Merozoite Invasion and Regulates Artemisinin Sensitivity
Abstract
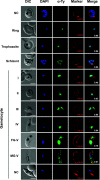
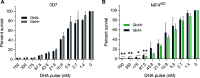
Artemisinin resistance in Plasmodium falciparum has been associated with a mutation in the NLI-interacting factor-like phosphatase PfNIF4, in addition to the mutations in the Kelch13 protein as the major determinant. We found that PfNIF4 was predominantly expressed at the schizont stage and localized in the nuclei of the parasite. To elucidate the functions of PfNIF4 in P. falciparum, we performed PfNIF4 knockdown (KD) using the inducible ribozyme system. PfNIF4 KD attenuated merozoite invasion and affected gametocytogenesis. PfNIF4 KD parasites also showed significantly increased in vitro susceptibility to artemisinins. Transcriptomic and proteomic analysis revealed that PfNIF4 KD led to the downregulation of gene categories involved in invasion and artemisinin resistance (e.g., mitochondrial function, membrane, and Kelch13 interactome) at the trophozoite and/or schizont stage. Consistent with PfNIF4 being a protein phosphatase, PfNIF4 KD resulted in an overall upregulation of the phosphoproteome of infected erythrocytes. Quantitative phosphoproteomic profiling identified a set of PfNIF4-regulated phosphoproteins with functional similarity to FCP1 substrates, particularly proteins involved in chromatin organization and transcriptional regulation. Specifically, we observed increased phosphorylation of Ser2/5 of the tandem repeats in the C-terminal domain (CTD) of RNA polymerase II (RNAPII) upon PfNIF4 KD. Furthermore, using the TurboID-based proteomic approach, we identified that PfNIF4 interacted with the RNAPII components, AP2-domain transcription factors, and chromatin-modifiers and binders. These findings suggest that PfNIF4 may act as the RNAPII CTD phosphatase, regulating the expression of general and parasite-specific cellular pathways during the blood-stage development. IMPORTANCE Protein phosphorylation regulates a multitude of cellular processes. The eukaryotic FCP1 phosphatase acts as a CTD-phosphatase to critically balance the phosphorylation status of the CTD of the RNAPII, controlling the accurate execution of the transcription process. Here, we identified PfNIF4 as the FCP1-like phosphatase in P. falciparum. PfNIF4 KD specifically downregulated genes involved in merozoite invasion, resulting in the attenuation of this process. Consistent with the earlier finding of the association of PfNIF4 mutations with artemisinin resistance in Southeast Asian parasite populations, PfNIF4 KD significantly increased in vitro susceptibility to artemisinins. The regulation of these cellular processes in P. falciparum by PfNIF4 is likely realized through RNAPII-mediated transcription, because PfNIF4 was found to interact with RNAPII subunits and KD of PfNIF4 caused CTD hyperphosphorylation. Our results reveal the functions of the PfNIF4 phosphatase in controlling the transcription of invasion- and resistance-related genes in the malaria parasite.
Keywords: RNA polymerase II; asexual development; invasion; malaria; protein phosphatase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
The Plasmodium falciparum Artemisinin Susceptibility-Associated AP-2 Adaptin μ Subunit is Clathrin Independent and Essential for Schizont Maturation.mBio. 2020 Feb 25;11(1):e02918-19. doi: 10.1128/mBio.02918-19. mBio. 2020. PMID: 32098816 Free PMC article.
-
Plasmodium berghei K13 Mutations Mediate In Vivo Artemisinin Resistance That Is Reversed by Proteasome Inhibition.mBio. 2020 Nov 10;11(6):e02312-20. doi: 10.1128/mBio.02312-20. mBio. 2020. PMID: 33173001 Free PMC article.
-
Role of Plasmodium falciparum Kelch 13 Protein Mutations in P. falciparum Populations from Northeastern Myanmar in Mediating Artemisinin Resistance.mBio. 2020 Feb 25;11(1):e01134-19. doi: 10.1128/mBio.01134-19. mBio. 2020. PMID: 32098812 Free PMC article.
-
Artemisinin Action and Resistance in Plasmodium falciparum.Trends Parasitol. 2016 Sep;32(9):682-696. doi: 10.1016/j.pt.2016.05.010. Epub 2016 Jun 9. Trends Parasitol. 2016. PMID: 27289273 Free PMC article. Review.
-
Extensive differential protein phosphorylation as intraerythrocytic Plasmodium falciparum schizonts develop into extracellular invasive merozoites.Proteomics. 2015 Aug;15(15):2716-29. doi: 10.1002/pmic.201400508. Epub 2015 May 19. Proteomics. 2015. PMID: 25886026 Review.
Cited by
-
Plasmodium falciparum protein phosphatase PP7 is required for early ring-stage development.mBio. 2024 Nov 13;15(11):e0253924. doi: 10.1128/mbio.02539-24. Epub 2024 Oct 10. mBio. 2024. PMID: 39387582 Free PMC article.
-
Genome sequencing of Plasmodium malariae identifies continental segregation and mutations associated with reduced pyrimethamine susceptibility.Nat Commun. 2024 Dec 30;15(1):10779. doi: 10.1038/s41467-024-55102-3. Nat Commun. 2024. PMID: 39738025 Free PMC article.
-
An inner membrane complex protein IMC1g in Plasmodium berghei is involved in asexual stage schizogony and parasite transmission.mBio. 2025 Jan 8;16(1):e0265224. doi: 10.1128/mbio.02652-24. Epub 2024 Nov 22. mBio. 2025. PMID: 39576115 Free PMC article.
References
-
- WHO Organization. 2020. World malaria report 2020. Global Malaria Programme, World Health Organization.
-
- Alam MM, Solyakov L, Bottrill AR, Flueck C, Siddiqui FA, Singh S, Mistry S, Viskaduraki M, Lee K, Hopp CS, Chitnis CE, Doerig C, Moon RW, Green JL, Holder AA, Baker DA, Tobin AB. 2015. Phosphoproteomics reveals malaria parasite Protein Kinase G as a signalling hub regulating egress and invasion. Nat Commun 6:7285. doi:10.1038/ncomms8285. - DOI - PMC - PubMed
-
- Pease BN, Huttlin EL, Jedrychowski MP, Talevich E, Harmon J, Dillman T, Kannan N, Doerig C, Chakrabarti R, Gygi SP, Chakrabarti D. 2013. Global analysis of protein expression and phosphorylation of three stages of Plasmodium falciparum intraerythrocytic development. J Proteome Res 12:4028–4045. doi:10.1021/pr400394g. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

