Human Nasal Organoids Model SARS-CoV-2 Upper Respiratory Infection and Recapitulate the Differential Infectivity of Emerging Variants
- PMID: 35938726
- PMCID: PMC9426414
- DOI: 10.1128/mbio.01944-22
Human Nasal Organoids Model SARS-CoV-2 Upper Respiratory Infection and Recapitulate the Differential Infectivity of Emerging Variants
Abstract
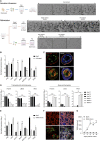
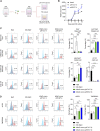
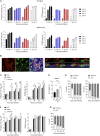
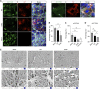
The human upper respiratory tract, specifically the nasopharyngeal epithelium, is the entry portal and primary infection site of respiratory viruses. Productive infection of SARS-CoV-2 in the nasal epithelium constitutes the cellular basis of viral pathogenesis and transmissibility. Yet a robust and well-characterized in vitro model of the nasal epithelium remained elusive. Here we report an organoid culture system of the nasal epithelium. We derived nasal organoids from easily accessible nasal epithelial cells with a perfect establishment rate. The derived nasal organoids were consecutively passaged for over 6 months. We then established differentiation protocols to generate 3-dimensional differentiated nasal organoids and organoid monolayers of 2-dimensional format that faithfully simulate the nasal epithelium. Moreover, when differentiated under a slightly acidic pH, the nasal organoid monolayers represented the optimal correlate of the native nasal epithelium for modeling the high infectivity of SARS-CoV-2, superior to all existing organoid models. Notably, the differentiated nasal organoid monolayers accurately recapitulated higher infectivity and replicative fitness of the Omicron variant than the prior variants. SARS-CoV-2, especially the more transmissible Delta and Omicron variants, destroyed ciliated cells and disassembled tight junctions, thereby facilitating virus spread and transmission. In conclusion, we establish a robust organoid culture system of the human nasal epithelium for modeling upper respiratory infections and provide a physiologically-relevant model for assessing the infectivity of SARS-CoV-2 emerging variants. IMPORTANCE An in vitro model of the nasal epithelium is imperative for understanding cell biology and virus-host interaction in the human upper respiratory tract. Here we report an organoid culture system of the nasal epithelium. Nasal organoids were derived from readily accessible nasal epithelial cells with perfect efficiency and stably expanded for more than 6 months. The long-term expandable nasal organoids were induced maturation into differentiated nasal organoids that morphologically and functionally simulate the nasal epithelium. The differentiated nasal organoids adequately recapitulated the higher infectivity and replicative fitness of SARS-CoV-2 emerging variants than the ancestral strain and revealed viral pathogenesis such as ciliary damage and tight junction disruption. Overall, we established a human nasal organoid culture system that enables a highly efficient reconstruction and stable expansion of the human nasal epithelium in culture plates, thus providing a facile and robust tool in the toolbox of microbiologists.
Keywords: SARS-CoV-2; airway organoids; ciliary damage; nasal organoids; tight junction.
Conflict of interest statement
The authors declare a conflict of interest. J.Z., K.Y.Y., M.C.C., and C.L. are listed as inventors on the patent of airway organoids (publication No: US-2021-0207081-A1). J.Z., M.C.C., C.L., and K.Y.Y. are listed as inventors on the provisional patent of nasal organoids (US application No: 63/358,795) describing the methods in this paper. All other authors declare no competing interests.
Figures
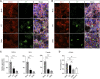
References
-
- Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. doi: 10.1016/S0140-6736(20)30154-9. - DOI - PMC - PubMed
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus I, Research T. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Ziegler CGK, Miao VN, Owings AH, Navia AW, Tang Y, Bromley JD, Lotfy P, Sloan M, Laird H, Williams HB, George M, Drake RS, Christian T, Parker A, Sindel CB, Burger MW, Pride Y, Hasan M, Abraham GE, 3rd, Senitko M, Robinson TO, Shalek AK, Glover SC, Horwitz BH, Ordovas-Montanes J. 2021. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell 184:4713–4733.e22. doi: 10.1016/j.cell.2021.07.023. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

