Impact of Bacillus subtilis Antibiotic Bacilysin and Campylobacter jejuni Efflux Pumps on Pathogen Survival in Mixed Biofilms
- PMID: 35938811
- PMCID: PMC9430781
- DOI: 10.1128/spectrum.02156-22
Impact of Bacillus subtilis Antibiotic Bacilysin and Campylobacter jejuni Efflux Pumps on Pathogen Survival in Mixed Biofilms
Abstract
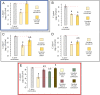
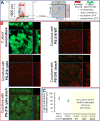
The foodborne pathogen Campylobacter jejuni is typically found in an agricultural environment; in animals, such as birds, as an intestinal commensal; and also in food products, especially fresh poultry meat. Campylobacter interactions within mixed species biofilms are poorly understood, especially at the microscale. We have recently shown that the beneficial bacterium Bacillus subtilis reduces C. jejuni survival and biofilm formation in coculture by secreting the antibiotic bacillaene. We extend these studies here by providing evidence that besides bacillaene, the antagonistic effect of B. subtilis involves a nonribosomal peptide bacilysin and that the fully functional antagonism depends on the quorum-sensing transcriptional regulator ComA. Using confocal laser scanning microscopy, we also show that secreted antibiotics influence the distribution of C. jejuni and B. subtilis cells in the submerged biofilm and decrease the thickness of the pathogen's biofilm. Furthermore, we demonstrate that genes encoding structural or regulatory proteins of the efflux apparatus system (cmeF and cmeR), respectively, contribute to the survival of C. jejuni during interaction with B. subtilis PS-216. In conclusion, this study demonstrates a strong potential of B. subtilis PS-216 to reduce C. jejuni biofilm growth, which supports the application of the PS-216 strain to pathogen biofilm control. IMPORTANCE Campylobacter jejuni is a prevalent cause of foodborne infections worldwide, while Bacillus subtilis as a potential probiotic represents an alternative strategy to control this alimentary infection. However, only limited literature exists on the specific mechanisms that shape interactions between B. subtilis and C. jejuni in biofilms. This study shows that in the two species biofilms, B. subtilis produces two antibiotics, bacillaene and bacilysin, that inhibit C. jejuni growth. In addition, we provide the first evidence that specific pathogen efflux pumps contribute to the defense against B. subtilis attack. Specifically, the CmeDEF pump acts during the defense against bacilysin, while CmeR-dependent overexpression of CmeABC nullifies the bacillaene attack. The role of specific B. subtilis antibiotics and these polyspecific pumps, known for providing resistance against medically relevant antibiotics, has not been studied during bacterial competition in biofilms before. Hence, this work broadens our understanding of mechanisms that shape antagonisms and defense during probiotic-pathogen interactions.
Keywords: Bacillus subtilis; Campylobacter jejuni; antibiotics; bacillaene; bacilysin; biofilm formation; efflux pumps; secondary metabolites.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Nutrient Availability and Biofilm Polysaccharide Shape the Bacillaene-Dependent Antagonism of Bacillus subtilis against Salmonella Typhimurium.Microbiol Spectr. 2022 Dec 21;10(6):e0183622. doi: 10.1128/spectrum.01836-22. Epub 2022 Nov 7. Microbiol Spectr. 2022. PMID: 36342318 Free PMC article.
-
Bacillaene Mediates the Inhibitory Effect of Bacillus subtilis on Campylobacter jejuni Biofilms.Appl Environ Microbiol. 2021 May 26;87(12):e0295520. doi: 10.1128/AEM.02955-20. Epub 2021 May 26. Appl Environ Microbiol. 2021. PMID: 33837012 Free PMC article.
-
Bacillus subtilis-derived peptides disrupt quorum sensing and biofilm assembly in multidrug-resistant Staphylococcus aureus.mSystems. 2024 Aug 20;9(8):e0071224. doi: 10.1128/msystems.00712-24. Epub 2024 Jul 11. mSystems. 2024. PMID: 38990088 Free PMC article.
-
Mechanistic concepts involved in biofilm associated processes of Campylobacter jejuni: persistence and inhibition in poultry environments.Poult Sci. 2024 Dec;103(12):104328. doi: 10.1016/j.psj.2024.104328. Epub 2024 Sep 12. Poult Sci. 2024. PMID: 39366290 Free PMC article. Review.
-
Resistance mechanisms in Campylobacter jejuni.Virulence. 2013 Apr 1;4(3):230-40. doi: 10.4161/viru.23753. Epub 2013 Feb 13. Virulence. 2013. PMID: 23406779 Free PMC article. Review.
Cited by
-
Important role of Bacillus subtilis as a probiotic and vaccine carrier in animal health maintenance.World J Microbiol Biotechnol. 2024 Jul 15;40(9):268. doi: 10.1007/s11274-024-04065-0. World J Microbiol Biotechnol. 2024. PMID: 39007987 Review.
-
Nutrient Availability and Biofilm Polysaccharide Shape the Bacillaene-Dependent Antagonism of Bacillus subtilis against Salmonella Typhimurium.Microbiol Spectr. 2022 Dec 21;10(6):e0183622. doi: 10.1128/spectrum.01836-22. Epub 2022 Nov 7. Microbiol Spectr. 2022. PMID: 36342318 Free PMC article.
-
Bacillus subtilis ensures high spore quality in competition with Salmonella Typhimurium via the SigB-dependent pathway.ISME J. 2025 Jan 2;19(1):wraf052. doi: 10.1093/ismejo/wraf052. ISME J. 2025. PMID: 40098255 Free PMC article.
-
A comprehensive description of the TolC effect on the antimicrobial susceptibility profile in Enterobacter bugandensis.Front Cell Infect Microbiol. 2022 Dec 9;12:1036933. doi: 10.3389/fcimb.2022.1036933. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36569193 Free PMC article.
-
Genomic Analysis and Metabolite Profiling of Three Probiotic Bacillus Strains for Potential Application in Aquaculture.Prev Nutr Food Sci. 2025 Jun 30;30(3):274-284. doi: 10.3746/pnf.2025.30.3.274. Prev Nutr Food Sci. 2025. PMID: 40612759 Free PMC article.
References
-
- WHO. 2020. Campylobacter. World Health Organization, Geneva, Switzerland. https://www.who.int/news-room/fact-sheets/detail/campylobacter.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

