AhrC Negatively Regulates Streptococcus mutans Arginine Biosynthesis
- PMID: 35938859
- PMCID: PMC9430756
- DOI: 10.1128/spectrum.00721-22
AhrC Negatively Regulates Streptococcus mutans Arginine Biosynthesis
Abstract
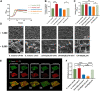
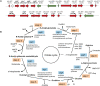
Streptococcus mutans is a primary cariogenic pathogen in humans. Arginine metabolism is required for bacterial growth. In S. mutans, however, the involvement of transcription factors in regulating arginine metabolism is unclear. The purpose of this study was to investigate the function and mechanism of ArgR family transcription factors in S. mutans. Here, we identified an ArgR (arginine repressor) family transcription factor named AhrC, which negatively regulates arginine biosynthesis and biofilm formation in S. mutans. The ahrC in-frame deletion strain exhibited slow growth and significantly increased intracellular arginine content. The strain overexpressing ahrC showed reduced intracellular arginine content, decreased biofilm biomass, reduced production of water-insoluble exopolysaccharides (EPS), and different biofilm structures. Furthermore, global gene expression profiles revealed differential expression levels of 233 genes in the ahrC-deficient strain, among which genes related to arginine biosynthesis (argJ, argB, argC, argD, argF, argG, argH) were significantly upregulated. In the ahrC overexpression strain, there are 89 differentially expressed genes, mostly related to arginine biosynthesis. The conserved DNA patterns bound by AhrC were identified by electrophoretic mobility shift assay (EMSA) and DNase I footprinting. In addition, the analysis of β-galactosidase activity showed that AhrC acted as a negative regulator. Taken together, our findings suggest that AhrC is an important transcription factor that regulates arginine biosynthesis gene expression and biofilm formation in S. mutans. These findings add new aspects to the complexity of regulating the expression of genes involved in arginine biosynthesis and biofilm formation in S. mutans. IMPORTANCE Arginine metabolism is essential for bacterial growth. The regulation of intracellular arginine metabolism in Streptococcus mutans, one of the major pathogens of dental caries, is unclear. In this study, we found that the transcription factor AhrC can directly and negatively regulate the expression of N-acetyl-gamma-glutamyl-phosphate reductase (argC), thus regulating arginine biosynthesis in S. mutans. In addition, the ahrC overexpression strain exhibited a significant decrease in biofilm and water-insoluble extracellular polysaccharides (EPS). This study adds new support to our understanding of the regulation of intracellular arginine metabolism in S. mutans.
Keywords: Streptococcus mutans; biofilm(s); gene expression; microbial genetics; transcription factor(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
EpsR Negatively Regulates Streptococcus mutans Exopolysaccharide Synthesis.J Dent Res. 2021 Aug;100(9):968-976. doi: 10.1177/00220345211000668. Epub 2021 Mar 20. J Dent Res. 2021. PMID: 33749354
-
ciaR impacts biofilm formation by regulating an arginine biosynthesis pathway in Streptococcus sanguinis SK36.Sci Rep. 2017 Dec 7;7(1):17183. doi: 10.1038/s41598-017-17383-1. Sci Rep. 2017. PMID: 29215019 Free PMC article.
-
Regulation of water-soluble glucan synthesis by the Streptococcus mutans dexA gene effects biofilm aggregation and cariogenic pathogenicity.Mol Oral Microbiol. 2019 Apr;34(2):51-63. doi: 10.1111/omi.12253. Epub 2019 Feb 14. Mol Oral Microbiol. 2019. PMID: 30659765
-
Inhibition of Streptococcus mutans biofilm formation by strategies targeting the metabolism of exopolysaccharides.Crit Rev Microbiol. 2021 Sep;47(5):667-677. doi: 10.1080/1040841X.2021.1915959. Epub 2021 May 3. Crit Rev Microbiol. 2021. PMID: 33938347 Review.
-
Polyketides/nonribosomal peptides from Streptococcus mutans and their ecological roles in dental biofilm.Mol Oral Microbiol. 2024 Oct;39(5):261-269. doi: 10.1111/omi.12451. Epub 2024 Jan 11. Mol Oral Microbiol. 2024. PMID: 38212261 Review.
Cited by
-
Saliva microbiome alterations in dental fluorosis population.J Oral Microbiol. 2023 Feb 20;15(1):2180927. doi: 10.1080/20002297.2023.2180927. eCollection 2023. J Oral Microbiol. 2023. PMID: 36844898 Free PMC article.
-
Proteomic study of the inhibitory effects of tannic acid on MRSA biofilm.Front Pharmacol. 2024 Dec 18;15:1413669. doi: 10.3389/fphar.2024.1413669. eCollection 2024. Front Pharmacol. 2024. PMID: 39744121 Free PMC article.
-
Glucosyltransferase activity-based screening identifies tannic acid as an inhibitor of Streptococcus mutans biofilm.Front Microbiol. 2025 Mar 27;16:1555497. doi: 10.3389/fmicb.2025.1555497. eCollection 2025. Front Microbiol. 2025. PMID: 40212388 Free PMC article.
-
Regulatory mechanisms of exopolysaccharide synthesis and biofilm formation in Streptococcus mutans.J Oral Microbiol. 2023 Jun 18;15(1):2225257. doi: 10.1080/20002297.2023.2225257. eCollection 2023. J Oral Microbiol. 2023. PMID: 37346997 Free PMC article. Review.
-
Dental caries prevalence and severity positively associate with AMY1 gene copy number.Clin Oral Investig. 2023 Dec 26;28(1):25. doi: 10.1007/s00784-023-05435-y. Clin Oral Investig. 2023. PMID: 38147184
References
-
- Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, Scisci EL, Hajishengallis E, Whiteley M, Koo H. 2020. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci USA 117:12375–12386. doi:10.1073/pnas.1919099117. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

