Helminth-induced reprogramming of the stem cell compartment inhibits type 2 immunity
- PMID: 35938990
- PMCID: PMC9365672
- DOI: 10.1084/jem.20212311
Helminth-induced reprogramming of the stem cell compartment inhibits type 2 immunity
Abstract
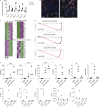
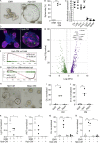
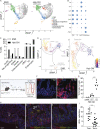
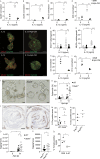
Enteric helminths form intimate physical connections with the intestinal epithelium, yet their ability to directly alter epithelial stem cell fate has not been resolved. Here we demonstrate that infection of mice with the parasite Heligmosomoides polygyrus bakeri (Hpb) reprograms the intestinal epithelium into a fetal-like state marked by the emergence of Clusterin-expressing revival stem cells (revSCs). Organoid-based studies using parasite-derived excretory-secretory products reveal that Hpb-mediated revSC generation occurs independently of host-derived immune signals and inhibits type 2 cytokine-driven differentiation of secretory epithelial lineages that promote their expulsion. Reciprocally, type 2 cytokine signals limit revSC differentiation and, consequently, Hpb fitness, indicating that helminths compete with their host for control of the intestinal stem cell compartment to promote continuation of their life cycle.
© 2022 Karo-Atar et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures




Comment in
-
Helminths revive to survive.J Exp Med. 2022 Sep 5;219(9):e20221183. doi: 10.1084/jem.20221183. Epub 2022 Aug 16. J Exp Med. 2022. PMID: 37559179 Free PMC article.
Similar articles
-
Production and analysis of immunomodulatory excretory-secretory products from the mouse gastrointestinal nematode Heligmosomoides polygyrus bakeri.Nat Protoc. 2014 Dec;9(12):2740-54. doi: 10.1038/nprot.2014.184. Epub 2014 Nov 6. Nat Protoc. 2014. PMID: 25375989
-
Novel O-linked methylated glycan antigens decorate secreted immunodominant glycoproteins from the intestinal nematode Heligmosomoides polygyrus.Int J Parasitol. 2016 Mar;46(3):157-170. doi: 10.1016/j.ijpara.2015.10.004. Epub 2015 Dec 12. Int J Parasitol. 2016. PMID: 26688390 Free PMC article.
-
Parasite Proximity Drives the Expansion of Regulatory T Cells in Peyer's Patches following Intestinal Helminth Infection.Infect Immun. 2015 Sep;83(9):3657-65. doi: 10.1128/IAI.00266-15. Epub 2015 Jul 6. Infect Immun. 2015. PMID: 26150538 Free PMC article.
-
Immune antibodies and helminth products drive CXCR2-dependent macrophage-myofibroblast crosstalk to promote intestinal repair.PLoS Pathog. 2015 Mar 25;11(3):e1004778. doi: 10.1371/journal.ppat.1004778. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25806513 Free PMC article.
-
Critical Role for Interleukin-25 in Host Protective Th2 Memory Response against Heligmosomoides polygyrus bakeri.Infect Immun. 2016 Nov 18;84(12):3328-3337. doi: 10.1128/IAI.00180-16. Print 2016 Dec. Infect Immun. 2016. PMID: 27620722 Free PMC article.
Cited by
-
Systemic Immune Modulation by Gastrointestinal Nematodes.Annu Rev Immunol. 2024 Jun;42(1):259-288. doi: 10.1146/annurev-immunol-090222-101331. Epub 2024 Jun 14. Annu Rev Immunol. 2024. PMID: 38277692 Free PMC article. Review.
-
Microbiome-based interventions to modulate gut ecology and the immune system.Mucosal Immunol. 2022 Jun;15(6):1095-1113. doi: 10.1038/s41385-022-00564-1. Epub 2022 Sep 30. Mucosal Immunol. 2022. PMID: 36180583 Free PMC article. Review.
-
Activation of fetal-like molecular programs during regeneration in the intestine and beyond.Cell Stem Cell. 2024 Jul 5;31(7):949-960. doi: 10.1016/j.stem.2024.05.009. Cell Stem Cell. 2024. PMID: 38971147 Free PMC article. Review.
-
Networking between helminths, microbes, and mammals.Cell Host Microbe. 2023 Apr 12;31(4):464-471. doi: 10.1016/j.chom.2023.02.008. Cell Host Microbe. 2023. PMID: 37054669 Free PMC article. Review.
-
Chromatin remodelling in damaged intestinal crypts orchestrates redundant TGFβ and Hippo signalling to drive regeneration.Nat Cell Biol. 2024 Dec;26(12):2084-2098. doi: 10.1038/s41556-024-01550-4. Epub 2024 Nov 15. Nat Cell Biol. 2024. PMID: 39548329
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

