Supercomputing Pipelines Search for Therapeutics Against COVID-19
- PMID: 35939280
- PMCID: PMC9280802
- DOI: 10.1109/MCSE.2020.3036540
Supercomputing Pipelines Search for Therapeutics Against COVID-19
Abstract
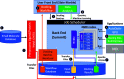
The urgent search for drugs to combat SARS-CoV-2 has included the use of supercomputers. The use of general-purpose graphical processing units (GPUs), massive parallelism, and new software for high-performance computing (HPC) has allowed researchers to search the vast chemical space of potential drugs faster than ever before. We developed a new drug discovery pipeline using the Summit supercomputer at Oak Ridge National Laboratory to help pioneer this effort, with new platforms that incorporate GPU-accelerated simulation and allow for the virtual screening of billions of potential drug compounds in days compared to weeks or months for their ability to inhibit SARS-COV-2 proteins. This effort will accelerate the process of developing drugs to combat the current COVID-19 pandemic and other diseases.
Figures




References
-
- Drews J., “Drug discovery: A historical perspective,” Science, vol. 287, no. 5460, pp. 1960–1964, 2000. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
