OverFlap PCR: A reliable approach for generating plasmid DNA libraries containing random sequences without a template bias
- PMID: 35939421
- PMCID: PMC9359533
- DOI: 10.1371/journal.pone.0262968
OverFlap PCR: A reliable approach for generating plasmid DNA libraries containing random sequences without a template bias
Abstract
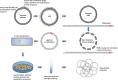
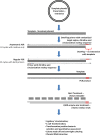
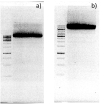
Over the decades, practical biotechnology researchers have aimed to improve naturally occurring proteins and create novel ones. It is widely recognized that coupling protein sequence randomization with various effect screening methodologies is one of the most powerful techniques for quickly, efficiently, and purposefully acquiring these desired improvements. Over the years, considerable advancements have been made in this field. However, developing PCR-based or template-guided methodologies has been hampered by resultant template sequence biases. Here, we present a novel whole plasmid amplification-based approach, which we named OverFlap PCR, for randomizing virtually any region of plasmid DNA without introducing a template sequence bias.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Thermostable DNA ligase-mediated PCR production of circular plasmid (PPCP) and its application in directed evolution via in situ error-prone PCR.DNA Res. 2013 Aug;20(4):375-82. doi: 10.1093/dnares/dst016. Epub 2013 Apr 30. DNA Res. 2013. PMID: 23633530 Free PMC article.
-
Random mutagenesis by whole-plasmid PCR amplification.Biotechniques. 1998 Mar;24(3):428-31. doi: 10.2144/98243st01. Biotechniques. 1998. PMID: 9526653
-
MEGAWHOP cloning: a method of creating random mutagenesis libraries via megaprimer PCR of whole plasmids.Methods Enzymol. 2011;498:399-406. doi: 10.1016/B978-0-12-385120-8.00017-6. Methods Enzymol. 2011. PMID: 21601687
-
Optimized PCR conditions minimizing the formation of chimeric DNA molecules from MPRA plasmid libraries.BMC Genomics. 2019 Jul 11;20(Suppl 7):536. doi: 10.1186/s12864-019-5847-2. BMC Genomics. 2019. PMID: 31291895 Free PMC article.
-
Polishing the craft of genetic diversity creation in directed evolution.Biotechnol Adv. 2013 Dec;31(8):1707-21. doi: 10.1016/j.biotechadv.2013.08.021. Epub 2013 Sep 6. Biotechnol Adv. 2013. PMID: 24012599 Review.
References
-
- Sarkar G, Sommer SS. The "megaprimer" method of site-directed mutagenesis. Biotechniques. 1990;8(4):404–7. . - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

