A comprehensive analysis of avian lymphoid leukosis-like lymphoma transcriptomes including identification of LncRNAs and the expression profiles
- PMID: 35939448
- PMCID: PMC9359530
- DOI: 10.1371/journal.pone.0272557
A comprehensive analysis of avian lymphoid leukosis-like lymphoma transcriptomes including identification of LncRNAs and the expression profiles
Abstract
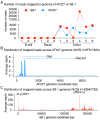
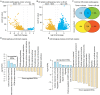
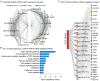
Avian lymphoid leukosis-like (LL-like) lymphoma has been observed in some experimental and commercial lines of chickens that are free of exogenous avian leukosis virus. Reported cases of avian lymphoid leukosis-like lymphoma incidences in the susceptible chickens are relatively low, but the apathogenic subgroup E avian leukosis virus (ALV-E) and the Marek's disease vaccine, SB-1, significantly escalate the disease incidence in the susceptible chickens. However, the underlying mechanism of tumorigenesis is poorly understood. In this study, we bioinformatically analyzed the deep RNA sequences of 6 lymphoid leukosis-like lymphoma samples, collected from susceptible chickens post both ALV-E and SB-1 inoculation, and identified a total of 1,692 novel long non-coding RNAs (lncRNAs). Thirty-nine of those novel lncRNAs were detected with altered expression in the LL-like tumors. In addition, 13 lncRNAs whose neighboring genes also showed differentially expression and 2 conserved novel lncRNAs, XLOC_001407 and XLOC_022595, may have previously un-appreciated roles in tumor development in human. Furthermore, 14 lncRNAs, especially XLOC_004542, exhibited strong potential as competing endogenous RNAs via sponging miRNAs. The analysis also showed that ALV subgroup E viral gene Gag/Gag-pol and the MD vaccine SB-1 viral gene R-LORF1 and ORF413 were particularly detectable in the LL-like tumor samples. In addition, we discovered 982 novel lncRNAs that were absent in the current annotation of chicken genome and 39 of them were aberrantly expressed in the tumors. This is the first time that lncRNA signature is identified in avian lymphoid leukosis-like lymphoma and suggests the epigenetic factor, lncRNA, is involved with the avian lymphoid leukosis-like lymphoma formation and development in susceptible chickens. Further studies to elucidate the genetic and epigenetic mechanisms underlying the avian lymphoid leukosis-like lymphoma is indeed warranted.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Endogenous Avian Leukosis Virus in Combination with Serotype 2 Marek's Disease Virus Significantly Boosted the Incidence of Lymphoid Leukosis-Like Bursal Lymphomas in Susceptible Chickens.J Virol. 2019 Nov 13;93(23):e00861-19. doi: 10.1128/JVI.00861-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31554689 Free PMC article.
-
Long non-coding RNA and MicroRNA profiling provides comprehensive insight into non-coding RNA involved host immune responses in ALV-J-infected chicken primary macrophage.Dev Comp Immunol. 2019 Nov;100:103414. doi: 10.1016/j.dci.2019.103414. Epub 2019 Jun 11. Dev Comp Immunol. 2019. PMID: 31200006
-
Augmentation of retrovirus-induced lymphoid leukosis by Marek's disease herpesviruses in White Leghorn chickens.J Virol. 1989 Feb;63(2):504-12. doi: 10.1128/JVI.63.2.504-512.1989. J Virol. 1989. PMID: 2536088 Free PMC article.
-
Discovery of novel long non-coding RNAs induced by subgroup J avian leukosis virus infection in chicken.Dev Comp Immunol. 2017 Nov;76:292-302. doi: 10.1016/j.dci.2017.06.015. Epub 2017 Jun 30. Dev Comp Immunol. 2017. PMID: 28673822
-
Isolation and characterization of an adventitious avian leukosis virus isolated from commercial Marek's disease vaccines.Avian Dis. 2006 Sep;50(3):380-5. doi: 10.1637/7497-122905R.1. Avian Dis. 2006. PMID: 17039837
References
-
- Cooper MD, Purchase HG, Bockman DE, Gathings WE. Studies on the nature of the abnormality of B cell differentiation in avian lymphoid leukosis: production of heterogeneous IgM by tumor cells. The Journal of Immunology. 1974;113(4):1210–22. - PubMed
-
- Purchase H. The pathogenesis and pathology of neoplasms caused by avian leukosis viruses. Avian leukosis: Springer; 1986. p. 171–96.
-
- Cooper M, Payne L, Dent P, Burmester B, Good R. Pathogenesis of Avian Lymphoid Leukosis. I. Histogenesis 2. Journal of the National Cancer Institute. 1968;41(2):373–89. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

