DNA binding by the Rad9A subunit of the Rad9-Rad1-Hus1 complex
- PMID: 35939452
- PMCID: PMC9359528
- DOI: 10.1371/journal.pone.0272645
DNA binding by the Rad9A subunit of the Rad9-Rad1-Hus1 complex
Abstract
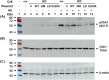
The Rad9-Rad1-Hus1 checkpoint clamp activates the DNA damage response and promotes DNA repair. DNA loading on the central channel of the Rad9-Rad1-Hus1 complex is required to execute its biological functions. Because Rad9A has the highest DNA affinity among the three subunits, we determined the domains and functional residues of human Rad9A that are critical for DNA interaction. The N-terminal globular domain (residues 1-133) had 3.7-fold better DNA binding affinity than the C-terminal globular domain (residues 134-266) of Rad9A1-266. Rad9A1-266 binds DNA 16-, 60-, and 30-fold better than Rad9A1-133, Rad9A134-266, and Rad9A94-266, respectively, indicating that different regions cooperatively contribute to DNA binding. We show that basic residues including K11, K15, R22, K78, K220, and R223 are important for DNA binding. The reductions on DNA binding of Ala substituted mutants of these basic residues show synergistic effect and are dependent on their residential Rad9A deletion constructs. Interestingly, deletion of a loop (residues 160-163) of Rad9A94-266 weakens DNA binding activity by 4.1-fold as compared to wild-type (WT) Rad9A94-266. Cellular sensitivity to genotoxin of rad9A knockout cells is restored by expressing WT-Rad9Afull. However, rad9A knockout cells expressing Rad9A mutants defective in DNA binding are more sensitive to H2O2 as compared to cells expressing WT-Rad9Afull. Only the rad9A knockout cells expressing loop-deleted Rad9A mutant are more sensitive to hydroxyurea than cells expressing WT-Rad9A. In addition, Rad9A-DNA interaction is required for DNA damage signaling activation. Our results indicate that DNA association by Rad9A is critical for maintaining cell viability and checkpoint activation under stress.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
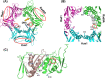




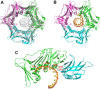
References
-
- Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, et al.. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A. 2003;100(4):1633–8. doi: 10.1073/pnas.0437927100 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

