Structural basis of higher order oligomerization of KSHV inhibitor of cGAS
- PMID: 35939686
- PMCID: PMC9388135
- DOI: 10.1073/pnas.2200285119
Structural basis of higher order oligomerization of KSHV inhibitor of cGAS
Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) inhibitor of cyclic GMP-AMP synthase (cGAS) (KicGAS) encoded by ORF52 is a conserved major tegument protein of KSHV and the first reported viral inhibitor of cGAS. In our previous study, we found that KicGAS is highly oligomerized in solution and that oligomerization is required for its cooperative DNA binding and for inhibiting DNA-induced phase separation and activation of cGAS. However, how KicGAS oligomerizes remained unclear. Here, we present the crystal structure of KicGAS at 2.5 Å resolution, which reveals an "L"-shaped molecule with each arm of the L essentially formed by a single α helix (α1 and α2). Antiparallel dimerization of α2 helices from two KicGAS molecules leads to a unique "Z"-shaped dimer. Surprisingly, α1 is also a dimerization domain. It forms a parallel dimeric leucine zipper with the α1 from a neighboring dimer, leading to the formation of an infinite chain of KicGAS dimers. Residues involved in leucine zipper dimer formation are among the most conserved residues across ORF52 homologs of gammaherpesviruses. The self-oligomerization increases the valence and cooperativity of interaction with DNA. The resultant multivalent interaction is critical for the formation of liquid condensates with DNA and consequent sequestration of DNA from being sensed by cGAS, explaining its role in restricting cGAS activation. The structure presented here not only provides a mechanistic understanding of the function of KicGAS but also informs a molecular target for rational design of antivirals against KSHV and related viruses.
Keywords: DNA binding; KicGAS; ORF52; cGAS; higher order oligomerization.
Conflict of interest statement
The authors declare no competing interest.
Figures
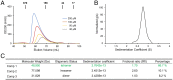


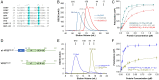

Similar articles
-
Cooperative DNA binding mediated by KicGAS/ORF52 oligomerization allows inhibition of DNA-induced phase separation and activation of cGAS.Nucleic Acids Res. 2021 Sep 20;49(16):9389-9403. doi: 10.1093/nar/gkab689. Nucleic Acids Res. 2021. PMID: 34387695 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Inhibitor of cGAS (KicGAS), Encoded by ORF52, Is an Abundant Tegument Protein and Is Required for Production of Infectious Progeny Viruses.J Virol. 2016 May 12;90(11):5329-5342. doi: 10.1128/JVI.02675-15. Print 2016 Jun 1. J Virol. 2016. PMID: 27009954 Free PMC article.
-
Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein.Cell Host Microbe. 2015 Sep 9;18(3):333-44. doi: 10.1016/j.chom.2015.07.015. Epub 2015 Aug 27. Cell Host Microbe. 2015. PMID: 26320998 Free PMC article.
-
KSHV strategies for host dsDNA sensing machinery.Virol Sin. 2016 Dec;31(6):466-471. doi: 10.1007/s12250-016-3877-3. Epub 2016 Dec 5. Virol Sin. 2016. PMID: 27933565 Free PMC article. Review.
-
Molecular mechanisms and cellular functions of cGAS-STING signalling.Nat Rev Mol Cell Biol. 2020 Sep;21(9):501-521. doi: 10.1038/s41580-020-0244-x. Epub 2020 May 18. Nat Rev Mol Cell Biol. 2020. PMID: 32424334 Review.
Cited by
-
Biomolecular phase separation in stress granule assembly and virus infection.Acta Biochim Biophys Sin (Shanghai). 2023 Jul 3;55(7):1099-1118. doi: 10.3724/abbs.2023117. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37401177 Free PMC article.
-
Viral evasion of the interferon response at a glance.J Cell Sci. 2023 Jun 15;136(12):jcs260682. doi: 10.1242/jcs.260682. Epub 2023 Jun 21. J Cell Sci. 2023. PMID: 37341132 Free PMC article. Review.
-
cGAMP-activated cGAS-STING signaling: its bacterial origins and evolutionary adaptation by metazoans.Nat Struct Mol Biol. 2023 Mar;30(3):245-260. doi: 10.1038/s41594-023-00933-9. Epub 2023 Mar 9. Nat Struct Mol Biol. 2023. PMID: 36894694 Free PMC article. Review.
-
Induction of translation-suppressive G3BP1+ stress granules and interferon-signaling cGAS condensates by transfected plasmid DNA.Hlife. 2025 Jan;3(1):21-37. doi: 10.1016/j.hlife.2024.11.005. Epub 2024 Dec 24. Hlife. 2025. PMID: 40078969 Free PMC article.
-
Pellino Proteins in Viral Immunity and Pathogenesis.Viruses. 2023 Jun 23;15(7):1422. doi: 10.3390/v15071422. Viruses. 2023. PMID: 37515108 Free PMC article. Review.
References
-
- Hopfner K.-P., Hornung V., Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AI146330/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 DE026101/DE/NIDCR NIH HHS/United States
- R01DE026101/HHS | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- R01 AI146330/AI/NIAID NIH HHS/United States
- R00AI108793/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources

