A nuclear redox sensor modulates gene activation and var switching in Plasmodium falciparum
- PMID: 35939693
- PMCID: PMC9388093
- DOI: 10.1073/pnas.2201247119
A nuclear redox sensor modulates gene activation and var switching in Plasmodium falciparum
Abstract
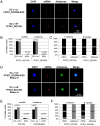
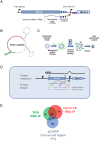
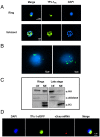
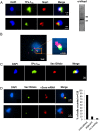
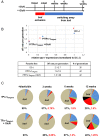
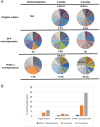
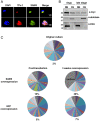
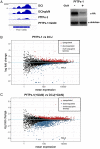
The virulence of Plasmodium falciparum, which causes the deadliest form of human malaria, is attributed to its ability to evade the human immune response. These parasites "choose" to express a single variant from a repertoire of surface antigens called PfEMP1, which are placed on the surface of the infected red cell. Immune evasion is achieved by switches in expression between var genes, each encoding a different PfEMP1 variant. While the mechanisms that regulate mutually exclusive expression of var genes are still elusive, antisense long-noncoding RNAs (lncRNAs) transcribed from the intron of the active var gene were implicated in the "choice" of the single active var gene. Here, we show that this lncRNA colocalizes with the site of var mRNA transcription and is anchored to the var locus via DNA:RNA interactions. We define the var lncRNA interactome and identify a redox sensor, P. falciparum thioredoxin peroxidase I (PfTPx-1), as one of the proteins associated with the var antisense lncRNA. We show that PfTPx-1 localizes to a nuclear subcompartment associated with active transcription on the nuclear periphery, in ring-stage parasite, when var transcription occurs. In addition, PfTPx-1 colocalizes with S-adenosylmethionine synthetase (PfSAMS) in the nucleus, and its overexpression leads to activation of var2csa, similar to overexpression of PfSAMS. Furthermore, we show that PfTPx-1 knockdown alters the var switch rate as well as activation of additional gene subsets. Taken together, our data indicate that nuclear PfTPx-1 plays a role in gene activation possibly by providing a redox-controlled nuclear microenvironment ideal for active transcription.
Keywords: Plasmodium falciparum; lncRNA; malaria; thioredoxin peroxidase; var genes.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
CRISPR/Cas9 Genome Editing Reveals That the Intron Is Not Essential for var2csa Gene Activation or Silencing in Plasmodium falciparum.mBio. 2017 Jul 11;8(4):e00729-17. doi: 10.1128/mBio.00729-17. mBio. 2017. PMID: 28698275 Free PMC article.
-
CRISPR Interference of a Clonally Variant GC-Rich Noncoding RNA Family Leads to General Repression of var Genes in Plasmodium falciparum.mBio. 2020 Jan 21;11(1):e03054-19. doi: 10.1128/mBio.03054-19. mBio. 2020. PMID: 31964736 Free PMC article.
-
A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite Plasmodium falciparum.PLoS Genet. 2015 May 19;11(5):e1005234. doi: 10.1371/journal.pgen.1005234. eCollection 2015 May. PLoS Genet. 2015. PMID: 25993442 Free PMC article.
-
PfEMP1: an antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum.Int J Biochem Cell Biol. 2009 Jul;41(7):1463-6. doi: 10.1016/j.biocel.2008.12.012. Epub 2008 Dec 25. Int J Biochem Cell Biol. 2009. PMID: 19150410 Review.
-
A greedy promoter controls malarial var-iations.Cell. 2006 Jan 27;124(2):251-3. doi: 10.1016/j.cell.2006.01.004. Cell. 2006. PMID: 16439197 Review.
Cited by
-
Variable surface antigen expression, virulence, and persistent infection by Plasmodium falciparum malaria parasites.Microbiol Mol Biol Rev. 2025 Mar 27;89(1):e0011423. doi: 10.1128/mmbr.00114-23. Epub 2025 Jan 14. Microbiol Mol Biol Rev. 2025. PMID: 39807932 Review.
-
PfEMP1 and var genes - Still of key importance in Plasmodium falciparum malaria pathogenesis and immunity.Adv Parasitol. 2024;125:53-103. doi: 10.1016/bs.apar.2024.02.001. Epub 2024 Mar 23. Adv Parasitol. 2024. PMID: 39095112 Free PMC article. Review.
-
Epigenetic Regulation and Chromatin Remodeling in Malaria Parasites.Annu Rev Microbiol. 2023 Sep 15;77:255-276. doi: 10.1146/annurev-micro-032521-041554. Epub 2023 Jun 2. Annu Rev Microbiol. 2023. PMID: 37268002 Free PMC article. Review.
-
SUN-domain proteins of the malaria parasite Plasmodium falciparum are essential for proper nuclear division and DNA repair.mBio. 2025 Apr 9;16(4):e0021625. doi: 10.1128/mbio.00216-25. Epub 2025 Mar 5. mBio. 2025. PMID: 40042312 Free PMC article.
-
Host-parasite interactions during Plasmodium infection: Implications for immunotherapies.Front Immunol. 2023 Jan 4;13:1091961. doi: 10.3389/fimmu.2022.1091961. eCollection 2022. Front Immunol. 2023. PMID: 36685595 Free PMC article. Review.
References
-
- (WHO) World Malaria Report 2020 (2020). https://www.who.int/publications/i/item/9789240015791. Accessed 31 July 2022.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

