Cohesin ATPase activities regulate DNA binding and coiled-coil configuration
- PMID: 35939705
- PMCID: PMC9388089
- DOI: 10.1073/pnas.2208004119
Cohesin ATPase activities regulate DNA binding and coiled-coil configuration
Abstract
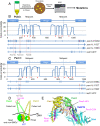
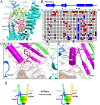
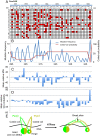
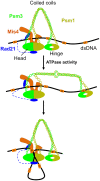
The cohesin complex is required for sister chromatid cohesion and genome compaction. Cohesin coiled coils (CCs) can fold at break sites near midpoints to bring head and hinge domains, located at opposite ends of coiled coils, into proximity. Whether ATPase activities in the head play a role in this conformational change is yet to be known. Here, we dissected functions of cohesin ATPase activities in cohesin dynamics in Schizosaccharomyces pombe. Isolation and characterization of cohesin ATPase temperature-sensitive (ts) mutants indicate that both ATPase domains are required for proper chromosome segregation. Unbiased screening of spontaneous suppressor mutations rescuing the temperature lethality of cohesin ATPase mutants identified several suppressor hotspots in cohesin that located outside of ATPase domains. Then, we performed comprehensive saturation mutagenesis targeted to these suppressor hotspots. Large numbers of the identified suppressor mutations indicated several different ways to compensate for the ATPase mutants: 1) Substitutions to amino acids with smaller side chains in coiled coils at break sites around midpoints may enable folding and extension of coiled coils more easily; 2) substitutions to arginine in the DNA binding region of the head may enhance DNA binding; or 3) substitutions to hydrophobic amino acids in coiled coils, connecting the head and interacting with other subunits, may alter conformation of coiled coils close to the head. These results reflect serial structural changes in cohesin driven by its ATPase activities potentially for packaging DNAs.
Keywords: ATPase; DNA binding; cohesin; coiled coil; suppressor screen.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Uhlmann F., SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 (2016). - PubMed
-
- Yatskevich S., Rhodes J., Nasmyth K., Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet. 53, 445–482 (2019). - PubMed
-
- Arumugam P., et al. , ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 13, 1941–1953 (2003). - PubMed
-
- Haering C. H., Löwe J., Hochwagen A., Nasmyth K., Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 (2002). - PubMed
-
- Lammens A., Schele A., Hopfner K. P., Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr. Biol. 14, 1778–1782 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

