DOCK2 is involved in the host genetics and biology of severe COVID-19
- PMID: 35940203
- PMCID: PMC9492544
- DOI: 10.1038/s41586-022-05163-5
DOCK2 is involved in the host genetics and biology of severe COVID-19
Abstract
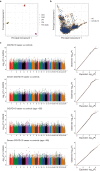
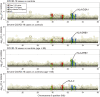
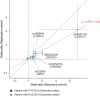
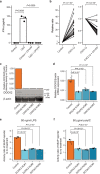
Identifying the host genetic factors underlying severe COVID-19 is an emerging challenge1-5. Here we conducted a genome-wide association study (GWAS) involving 2,393 cases of COVID-19 in a cohort of Japanese individuals collected during the initial waves of the pandemic, with 3,289 unaffected controls. We identified a variant on chromosome 5 at 5q35 (rs60200309-A), close to the dedicator of cytokinesis 2 gene (DOCK2), which was associated with severe COVID-19 in patients less than 65 years of age. This risk allele was prevalent in East Asian individuals but rare in Europeans, highlighting the value of genome-wide association studies in non-European populations. RNA-sequencing analysis of 473 bulk peripheral blood samples identified decreased expression of DOCK2 associated with the risk allele in these younger patients. DOCK2 expression was suppressed in patients with severe cases of COVID-19. Single-cell RNA-sequencing analysis (n = 61 individuals) identified cell-type-specific downregulation of DOCK2 and a COVID-19-specific decreasing effect of the risk allele on DOCK2 expression in non-classical monocytes. Immunohistochemistry of lung specimens from patients with severe COVID-19 pneumonia showed suppressed DOCK2 expression. Moreover, inhibition of DOCK2 function with CPYPP increased the severity of pneumonia in a Syrian hamster model of SARS-CoV-2 infection, characterized by weight loss, lung oedema, enhanced viral loads, impaired macrophage recruitment and dysregulated type I interferon responses. We conclude that DOCK2 has an important role in the host immune response to SARS-CoV-2 infection and the development of severe COVID-19, and could be further explored as a potential biomarker and/or therapeutic target.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

