Expression of CYP450 enzymes in human fetal membranes and its implications in xenobiotic metabolism during pregnancy
- PMID: 35940219
- PMCID: PMC12358170
- DOI: 10.1016/j.lfs.2022.120867
Expression of CYP450 enzymes in human fetal membranes and its implications in xenobiotic metabolism during pregnancy
Abstract
Background: Environmental exposure to toxicants is a major risk factor for spontaneous preterm birth (PTB, <37 weeks). Toxicants and drugs administered to patients are metabolized primarily by the cytochrome P450 (CYP450) system. Along with the adult and fetal liver, the placenta, a critical feto-maternal interface organ, expresses CYP450 enzymes that metabolize these xenobiotics. However, the contribution of the fetal membranes, another tissue of the feto-maternal interface, to the expression of CYP450 enzymes and the detoxification of xenobiotics remains unknown.
Aims: This study characterized CYP450 expression and determined the functional activity of CYP450 enzymes in fetal membranes.
Main methods: RNA sequencing (RNA-Seq) of placental and fetal membrane tissues and cells was done. Differential expressions of CYP450 genes were compared and validated via reverse transcription-quantitative polymerase chain reaction (RT-qPCR) between the two tissues. The functional activity of major CYP450 enzymes was determined using a fluorophore-based enzymatic assay in the presence and absence of their corresponding inhibitors.
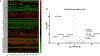
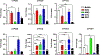
Key findings: With the exception of genes that regulate cholesterol metabolism, the expression profile of CYP450 genes was similar between placental and fetal membranes tissues/cells. RT-qPCR analysis confirmed these findings with significant levels of mRNA for major CYP450 genes being detectable in amnion epithelial cells (AECs) and chorion trophoblasts cells (CTCs). Biochemical analyses revealed significant CYP450 enzymatic activities that were sensitive to specific inhibitors for both AECs and CTCs, suggesting that the genes were expressed as functional enzymes.
Significance: This is the first study to determine global expression of CYP450 enzymes in fetal membranes which may play a role in xenobiotic metabolism during pregnancy. Given that many women are exposed to environmental toxins or require medications during pregnancy, a better understanding of their role in metabolism is required to develop safer therapeutics and prevent adverse outcomes.
Keywords: Cytochrome P450; Drug metabolism; Fetal membrane; Pharmacokinetics; Placenta; Pregnancy; Toxicant metabolism.
Copyright © 2022 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Use of biochemical tests of placental function for improving pregnancy outcome.Cochrane Database Syst Rev. 2015 Nov 25;2015(11):CD011202. doi: 10.1002/14651858.CD011202.pub2. Cochrane Database Syst Rev. 2015. PMID: 26602956 Free PMC article.
-
Placental Exosomes During Gestation: Liquid Biopsies Carrying Signals for the Regulation of Human Parturition.Curr Pharm Des. 2018;24(9):974-982. doi: 10.2174/1381612824666180125164429. Curr Pharm Des. 2018. PMID: 29376493 Free PMC article. Review.
-
Prenatal interventions for congenital diaphragmatic hernia for improving outcomes.Cochrane Database Syst Rev. 2015 Nov 27;2015(11):CD008925. doi: 10.1002/14651858.CD008925.pub2. Cochrane Database Syst Rev. 2015. PMID: 26611822 Free PMC article.
-
Different corticosteroids and regimens for accelerating fetal lung maturation for babies at risk of preterm birth.Cochrane Database Syst Rev. 2022 Aug 9;8(8):CD006764. doi: 10.1002/14651858.CD006764.pub4. Cochrane Database Syst Rev. 2022. PMID: 35943347 Free PMC article.
Cited by
-
Determining Sex-Specific Gene Expression Differences in Human Chorion Trophoblast Cells.Int J Mol Sci. 2025 Mar 2;26(5):2239. doi: 10.3390/ijms26052239. Int J Mol Sci. 2025. PMID: 40076861 Free PMC article.
-
Microfluidic technology and simulation models in studying pharmacokinetics during pregnancy.Front Pharmacol. 2023 Aug 17;14:1241815. doi: 10.3389/fphar.2023.1241815. eCollection 2023. Front Pharmacol. 2023. PMID: 37663251 Free PMC article.
-
Decoding the molecular pathways governing trophoblast migration and placental development; a literature review.Front Endocrinol (Lausanne). 2024 Nov 27;15:1486608. doi: 10.3389/fendo.2024.1486608. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39665023 Free PMC article. Review.
-
High-Throughput 3D-Printed Model of the Feto-Maternal Interface for the Discovery and Development of Preterm Birth Therapies.ACS Appl Mater Interfaces. 2024 Aug 14;16(32):41892-41906. doi: 10.1021/acsami.4c08731. Epub 2024 Jul 30. ACS Appl Mater Interfaces. 2024. PMID: 39078878 Free PMC article.
-
Augmentation of Pectoral Fin Teratogenicity by Thalidomide in Human Cytochrome P450 3A-Expressing Zebrafish.Pharmaceuticals (Basel). 2023 Feb 28;16(3):368. doi: 10.3390/ph16030368. Pharmaceuticals (Basel). 2023. PMID: 36986467 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

