Endoplasmic Reticulum-Associated Protein Degradation
- PMID: 35940909
- PMCID: PMC9732900
- DOI: 10.1101/cshperspect.a041247
Endoplasmic Reticulum-Associated Protein Degradation
Abstract
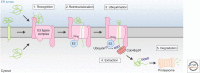
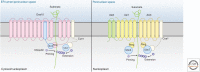
Misfolded, potentially toxic proteins in the lumen and membrane of the endoplasmic reticulum (ER) are eliminated by proteasomes in the cytosol through ER-associated degradation (ERAD). The ERAD process involves the recognition of substrates in the lumen and membrane of the ER, their translocation into the cytosol, ubiquitination, and delivery to the proteasome for degradation. These ERAD steps are performed by membrane-embedded ubiquitin-ligase complexes of different specificity that together cover a wide range of substrates. Besides misfolded proteins, ERAD further contributes to quality control by targeting unassembled and mislocalized proteins. ERAD also targets a restricted set of folded proteins to influence critical ER functions such as sterol biosynthesis, calcium homeostasis, or ER contacts with other organelles. This review describes the ubiquitin-ligase complexes and the principles guiding protein degradation by ERAD.
Copyright © 2022 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
