Adaptation to chronic ER stress enforces pancreatic β-cell plasticity
- PMID: 35941159
- PMCID: PMC9360004
- DOI: 10.1038/s41467-022-32425-7
Adaptation to chronic ER stress enforces pancreatic β-cell plasticity
Abstract
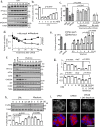
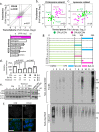
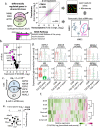
Pancreatic β-cells are prone to endoplasmic reticulum (ER) stress due to their role in insulin secretion. They require sustainable and efficient adaptive stress responses to cope with this stress. Whether episodes of chronic stress directly compromise β-cell identity is unknown. We show here under reversible, chronic stress conditions β-cells undergo transcriptional and translational reprogramming associated with impaired expression of regulators of β-cell function and identity. Upon recovery from stress, β-cells regain their identity and function, indicating a high degree of adaptive plasticity. Remarkably, while β-cells show resilience to episodic ER stress, when episodes exceed a threshold, β-cell identity is gradually lost. Single cell RNA-sequencing analysis of islets from type 1 diabetes patients indicates severe deregulation of the chronic stress-adaptation program and reveals novel biomarkers of diabetes progression. Our results suggest β-cell adaptive exhaustion contributes to diabetes pathogenesis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK053307/DK/NIDDK NIH HHS/United States
- U01 DK112217/DK/NIDDK NIH HHS/United States
- U01 DK127747/DK/NIDDK NIH HHS/United States
- R56 DK128136/DK/NIDDK NIH HHS/United States
- R37 DK060596/DK/NIDDK NIH HHS/United States
- UC4 DK112232/DK/NIDDK NIH HHS/United States
- R01 DK060596/DK/NIDDK NIH HHS/United States
- R01 CA217992/CA/NCI NIH HHS/United States
- U01 DK123594/DK/NIDDK NIH HHS/United States
- UC4 DK112217/DK/NIDDK NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- U01 DK123716/DK/NIDDK NIH HHS/United States
- R01 DK111174/DK/NIDDK NIH HHS/United States
- UC4 DK104155/DK/NIDDK NIH HHS/United States
- R01 DK130919/DK/NIDDK NIH HHS/United States
- R01 DK048280/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

