An updated review of SARS-CoV-2 detection methods in the context of a novel coronavirus pandemic
- PMID: 35942232
- PMCID: PMC9349698
- DOI: 10.1002/btm2.10356
An updated review of SARS-CoV-2 detection methods in the context of a novel coronavirus pandemic
Abstract
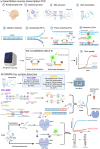
The World Health Organization has reported approximately 430 million confirmed cases of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), worldwide, including nearly 6 million deaths, since its initial appearance in China in 2019. While the number of diagnosed cases continues to increase, the need for technologies that can accurately and rapidly detect SARS-CoV-2 virus infection at early phases continues to grow, and the Federal Drug Administration (FDA) has licensed emergency use authorizations (EUAs) for virtually hundreds of diagnostic tests based on nucleic acid molecules and antigen-antibody serology assays. Among them, the quantitative real-time reverse transcription PCR (qRT-PCR) assay is considered the gold standard for early phase virus detection. Unfortunately, qRT-PCR still suffers from disadvantages such as the complex test process and the occurrence of false negatives; therefore, new nucleic acid detection devices and serological testing technologies are being developed. However, because of the emergence of strongly infectious mutants of the new coronavirus, such as Alpha (B.1.1.7), Delta (B.1.617.2), and Omicron (B.1.1.529), the need for the specific detection of mutant strains is also increasing. Therefore, this article reviews nucleic acid- and antigen-antibody-based serological assays, and compares the performance of some of the most recent FDA-approved and literature-reported assays and associated kits for the specific testing of new coronavirus variants.
Keywords: SARS‐CoV‐2; nucleic acid molecular test; serological test; test kit evaluation; viral variants; virus detection.
© 2022 The Authors. Bioengineering & Translational Medicine published by Wiley Periodicals LLC on behalf of American Institute of Chemical Engineers.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

