Sulforaphane, a secondary metabolite in crucifers, inhibits the oxidative stress adaptation and virulence of Xanthomonas by directly targeting OxyR
- PMID: 35942507
- PMCID: PMC9452769
- DOI: 10.1111/mpp.13245
Sulforaphane, a secondary metabolite in crucifers, inhibits the oxidative stress adaptation and virulence of Xanthomonas by directly targeting OxyR
Abstract
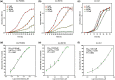
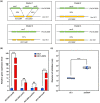
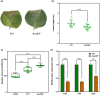
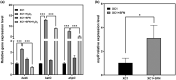
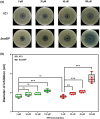
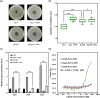
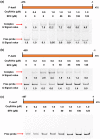
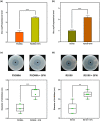
Plant secondary metabolites perform numerous functions in the interactions between plants and pathogens. However, little is known about the precise mechanisms underlying their contribution to the direct inhibition of pathogen growth and virulence in planta. Here, we show that the secondary metabolite sulforaphane (SFN) in crucifers inhibits the growth, virulence, and ability of Xanthomonas species to adapt to oxidative stress, which is essential for the successful infection of host plants by phytopathogens. The transcription of oxidative stress detoxification-related genes (catalase [katA and katG] and alkylhydroperoxide-NADPH oxidoreductase subunit C [ahpC]) was substantially inhibited by SFN in Xanthomonas campestris pv. campestris (Xcc), and this phenomenon was most obvious in sax gene mutants sensitive to SFN. By performing microscale thermophoresis (MST) and electrophoretic mobility shift assay (EMSA), we observed that SFN directly bound to the virulence-related redox-sensing transcription factor OxyR and weakened the ability of OxyR to bind to the promoters of oxidative stress detoxification-related genes. Collectively, these results illustrate that SFN directly targets OxyR to inhibit the bacterial adaptation to oxidative stress, thereby decreasing bacterial virulence. Interestingly, this phenomenon occurs in multiple Xanthomonas species. This study provides novel insights into the molecular mechanisms by which SFN limits Xanthomonas adaptation to oxidative stress and virulence, and the findings will facilitate future studies on the use of SFN as a biopesticide to control Xanthomonas.
Keywords: OxyR; Xanthomonas species; oxidative stress; secondary metabolites; sulforaphane; virulence.
© 2022 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures
Similar articles
-
SstF, a novel sulforaphane-sensing transcription factor of Xanthomonas campestris, is required for sulforaphane tolerance and virulence.Mol Plant Pathol. 2023 May;24(5):452-465. doi: 10.1111/mpp.13314. Epub 2023 Feb 24. Mol Plant Pathol. 2023. PMID: 36829260 Free PMC article.
-
Evaluation of the virulence of Xanthomonas campestris pv. campestris mutant strains lacking functional genes in the OxyR regulon.Curr Microbiol. 2011 Aug;63(2):232-7. doi: 10.1007/s00284-011-9970-9. Epub 2011 Jun 28. Curr Microbiol. 2011. PMID: 21710133
-
OxyR-regulated catalase CatB promotes the virulence in rice via detoxifying hydrogen peroxide in Xanthomonas oryzae pv. oryzae.BMC Microbiol. 2016 Nov 8;16(1):269. doi: 10.1186/s12866-016-0887-0. BMC Microbiol. 2016. PMID: 27825304 Free PMC article.
-
OxyR mediated compensatory expression between ahpC and katA and the significance of ahpC in protection from hydrogen peroxide in Xanthomonas campestris.FEMS Microbiol Lett. 2005 Aug 1;249(1):73-8. doi: 10.1016/j.femsle.2005.06.002. FEMS Microbiol Lett. 2005. PMID: 15993009
-
Genes for hydrogen peroxide detoxification and adaptation contribute to protection against heat shock in Xanthomonas campestris pv. campestris.FEMS Microbiol Lett. 2011 Apr;317(1):60-6. doi: 10.1111/j.1574-6968.2011.02211.x. Epub 2011 Jan 31. FEMS Microbiol Lett. 2011. PMID: 21219417
Cited by
-
SstF, a novel sulforaphane-sensing transcription factor of Xanthomonas campestris, is required for sulforaphane tolerance and virulence.Mol Plant Pathol. 2023 May;24(5):452-465. doi: 10.1111/mpp.13314. Epub 2023 Feb 24. Mol Plant Pathol. 2023. PMID: 36829260 Free PMC article.
-
How plants manage pathogen infection.EMBO Rep. 2024 Jan;25(1):31-44. doi: 10.1038/s44319-023-00023-3. Epub 2023 Dec 19. EMBO Rep. 2024. PMID: 38177909 Free PMC article. Review.
-
Iron ions regulate antifungal HSAF biosynthesis in Lysobacter enzymogenes by manipulating the DNA-binding affinity of the ferric uptake regulator (Fur).Microbiol Spectr. 2023 Sep 22;11(5):e0061723. doi: 10.1128/spectrum.00617-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37737630 Free PMC article.
-
Bioengineering for the Microbial Degradation of Petroleum Hydrocarbon Contaminants.Bioengineering (Basel). 2023 Mar 10;10(3):347. doi: 10.3390/bioengineering10030347. Bioengineering (Basel). 2023. PMID: 36978738 Free PMC article. Review.
-
OxyR is required for oxidative stress resistance of the entomopathogenic bacterium Xenorhabdus nematophila and has a minor role during the bacterial interaction with its hosts.Microbiology (Reading). 2024 Jul;170(7):001481. doi: 10.1099/mic.0.001481. Microbiology (Reading). 2024. PMID: 39058385 Free PMC article.
References
-
- Ahuja, I. , Kissen, R. & Bones, A.M. (2012) Phytoalexins in defense against pathogens. Trends in Plant Science, 17, 73–90. - PubMed
-
- Büttner, D. & Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiology Reviews, 34, 107–133. - PubMed
-
- Cabiscol, E. , Tamarit, J. & Ros, J. (2000) Oxidative stress in bacteria and protein damage by reactive oxygen species. International Journal of Microbiology, 3, 3–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

