Proton Motive Force Inhibitors Are Detrimental to Methicillin-Resistant Staphylococcus aureus Strains
- PMID: 35943153
- PMCID: PMC9430991
- DOI: 10.1128/spectrum.02024-22
Proton Motive Force Inhibitors Are Detrimental to Methicillin-Resistant Staphylococcus aureus Strains
Abstract
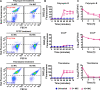
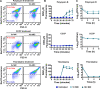
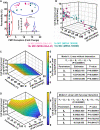
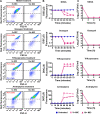
Methicillin-resistant Staphylococcus aureus (MRSA) strains are tolerant of conventional antibiotics, making them extremely dangerous. Previous studies have shown the effectiveness of proton motive force (PMF) inhibitors at killing bacterial cells; however, whether these agents can launch a new treatment strategy to eliminate antibiotic-tolerant cells mandates further investigation. Here, using known PMF inhibitors and two different MRSA isolates, we showed that the bactericidal potency of PMF inhibitors seemed to correlate with their ability to disrupt PMF and permeabilize cell membranes. By screening a small chemical library to verify this correlation, we identified a subset of chemicals (including nordihydroguaiaretic acid, gossypol, trifluoperazine, and amitriptyline) that strongly disrupted PMF in MRSA cells by dissipating either the transmembrane electric potential (ΔΨ) or the proton gradient (ΔpH). These drugs robustly permeabilized cell membranes and reduced MRSA cell levels below the limit of detection. Overall, our study further highlights the importance of cellular PMF as a target for designing new bactericidal therapeutics for pathogens. IMPORTANCE Methicillin-resistant Staphylococcus aureus (MRSA) emerged as a major hypervirulent pathogen that causes severe health care-acquired infections. These pathogens can be multidrug-tolerant cells, which can facilitate the recurrence of chronic infections and the emergence of diverse antibiotic-resistant mutants. In this study, we aimed to investigate whether proton motive force (PMF) inhibitors can launch a new treatment strategy to eliminate MRSA cells. Our in-depth analysis showed that PMF inhibitors that strongly dissipate either the transmembrane electric potential or the proton gradient can robustly permeabilize cell membranes and reduce MRSA cell levels below the limit of detection.
Keywords: PMF inhibitors; high-throughput drug screening; membrane permeabilization; methicillin-resistant Staphylococcus aureus; proton motive force; tolerant cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Natural Flavones from Morus alba against Methicillin-Resistant Staphylococcus aureus via Targeting the Proton Motive Force and Membrane Permeability.J Agric Food Chem. 2019 Sep 11;67(36):10222-10234. doi: 10.1021/acs.jafc.9b01795. Epub 2019 Sep 3. J Agric Food Chem. 2019. PMID: 31385700
-
Collapsing the proton motive force to identify synergistic combinations against Staphylococcus aureus.Chem Biol. 2013 Sep 19;20(9):1168-78. doi: 10.1016/j.chembiol.2013.07.006. Epub 2013 Aug 22. Chem Biol. 2013. PMID: 23972939
-
Isoniazid potentiates tigecycline to kill methicillin-resistant Staphylococcus aureus.Emerg Microbes Infect. 2025 Dec;14(1):2434587. doi: 10.1080/22221751.2024.2434587. Epub 2024 Dec 9. Emerg Microbes Infect. 2025. PMID: 39585340 Free PMC article.
-
Bacterial proton motive force as an unprecedented target to control antimicrobial resistance.Med Res Rev. 2023 Jul;43(4):1068-1090. doi: 10.1002/med.21946. Epub 2023 Mar 10. Med Res Rev. 2023. PMID: 36896761 Review.
-
Correlation Between Biofilm Formation and Antibiotic Resistance in MRSA and MSSA Isolated from Clinical Samples in Iran: A Systematic Review and Meta-Analysis.Microb Drug Resist. 2020 Sep;26(9):1071-1080. doi: 10.1089/mdr.2020.0001. Epub 2020 Mar 10. Microb Drug Resist. 2020. PMID: 32159447
Cited by
-
Reducing Peptidoglycan Crosslinking by Chemical Modulator Reverts β-lactam Resistance in Methicillin-Resistant Staphylococcus aureus.Adv Sci (Weinh). 2024 Jul;11(28):e2400858. doi: 10.1002/advs.202400858. Epub 2024 May 15. Adv Sci (Weinh). 2024. PMID: 38747156 Free PMC article.
-
Optimizing Niclosamide for Cancer Therapy: Improving Bioavailability via Structural Modification and Nanotechnology.Cancers (Basel). 2024 Oct 21;16(20):3548. doi: 10.3390/cancers16203548. Cancers (Basel). 2024. PMID: 39456642 Free PMC article. Review.
-
The Properties of Linezolid, Rifampicin, and Vancomycin, as Well as the Mechanism of Action of Pentamidine, Determine Their Synergy against Gram-Negative Bacteria.Int J Mol Sci. 2023 Sep 7;24(18):13812. doi: 10.3390/ijms241813812. Int J Mol Sci. 2023. PMID: 37762115 Free PMC article.
-
Unveiling the critical roles of cellular metabolism suppression in antibiotic tolerance.NPJ Antimicrob Resist. 2024 Jun 24;2(1):17. doi: 10.1038/s44259-024-00034-7. NPJ Antimicrob Resist. 2024. PMID: 39843626 Free PMC article.
-
Spatiotemporal dynamics of the proton motive force on single bacterial cells.Sci Adv. 2024 May 24;10(21):eadl5849. doi: 10.1126/sciadv.adl5849. Epub 2024 May 23. Sci Adv. 2024. PMID: 38781330 Free PMC article.
References
-
- Strauß L, Stegger M, Akpaka PE, Alabi A, Breurec S, Coombs G, Egyir B, Larsen AR, Laurent F, Monecke S, Peters G, Skov R, Strommenger B, Vandenesch F, Schaumburg F, Mellmann A. 2017. Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc Natl Acad Sci USA 114:E10596–E10604. doi:10.1073/pnas.1702472114. - DOI - PMC - PubMed
-
- Jevons MP. 1961. Celbenin-resistant staphylococci. Br Med J 1:124–125. doi:10.1136/bmj.1.5219.124-a. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

